Fas ligand
| View/Edit Human | View/Edit Mouse |
Fas ligand (FasL or CD95L) is a type-II transmembrane protein that belongs to the tumor necrosis factor (TNF) family. Its binding with its receptor induces apoptosis. Fas ligand/receptor interactions play an important role in the regulation of the immune system and the progression of cancer.
Structure
Fas ligand or FasL is a homotrimeric type II transmembrane protein expressed on cytotoxic T lymphocytes. It signals through trimerization of FasR, which spans the membrane of the "target" cell. This trimerization usually leads to apoptosis, or cell death.
Soluble Fas ligand is generated by cleaving membrane-bound FasL at a conserved cleavage site by the external matrix metalloproteinase MMP-7.
Receptors
- FasR: The Fas receptor (FasR), or CD95, is the most intensely studied member of the death receptor family. The gene is situated on chromosome 10 in humans and 19 in mice. Previous reports have identified as many as eight splice variants, which are translated into seven isoforms of the protein. Many of these isoforms are rare haplotypes that are usually associated with a state of disease. Apoptosis-inducing Fas receptor is dubbed isoform 1 and is a type 1 transmembrane protein. It consists of three cysteine-rich pseudorepeats, a transmembrane domain, and an intracellular death domain.
- DcR3: Decoy receptor 3 (DcR3) is a recently discovered decoy receptor of the tumor necrosis factor superfamily that binds to FasL, LIGHT, and TL1A. DcR3 is a soluble receptor that has no signal transduction capabilities (hence a "decoy") and functions to prevent FasR-FasL interactions by competitively binding to membrane-bound Fas ligand and rendering them inactive.[3]
Cell signaling
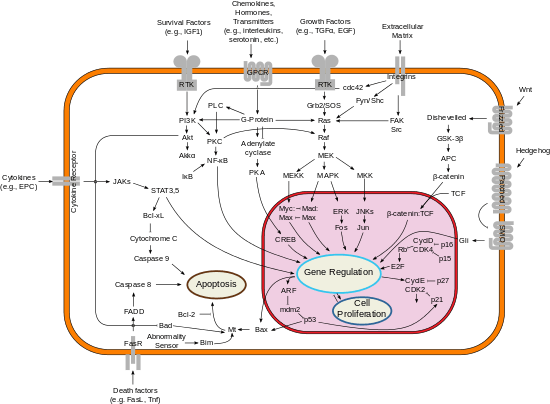
Fas forms the death-inducing signaling complex (DISC) upon ligand binding. Membrane-anchored Fas ligand trimer on the surface of an adjacent cell causes trimerization of Fas receptor. This event is also mimicked by binding of an agonistic Fas antibody, though some evidence suggests that the apoptotic signal induced by the antibody is unreliable in the study of Fas signaling. To this end, several clever ways of trimerizing the antibody for in vitro research have been employed.
Upon ensuing death domain (DD) aggregation, the receptor complex is internalized via the cellular endosomal machinery. This allows the adaptor molecule Fas-associated death domain (FADD) to bind the death domain of Fas through its own death domain. FADD also contains a death effector domain (DED) near its amino terminus, which facilitates binding to the DED of FADD-like ICE (FLICE), more commonly referred to as caspase-8. FLICE can then self-activate through proteolytic cleavage into p10 and p18 subunits, of which two form the active heterotetramer enzyme. Active caspase-8 is then released from the DISC into the cytosol, where it cleaves other effector caspases, eventually leading to DNA degradation, membrane blebbing, and other hallmarks of apoptosis.

Some reports have suggested that the extrinsic Fas pathway is sufficient to induce complete apoptosis in certain cell types through DISC assembly and subsequent caspase-8 activation. These cells are dubbed Type 1 cells and are characterized by the inability of anti-apoptotic members of the Bcl-2 family (namely Bcl-2 and Bcl-xL) to protect from Fas-mediated apoptosis. Characterized Type 1 cells include H9, CH1, SKW6.4, and SW480, all of which are lymphocyte lineages except the latter, which is of the colon adenocarcinoma lineage.
Evidence for crosstalk between the extrinsic and intrinsic pathways exists in the Fas signal cascade. In most cell types, caspase-8 catalyzes the cleavage of the pro-apoptotic BH3-only protein Bid into its truncated form, tBid. BH-3 only members of the Bcl-2 family engage exclusively anti-apoptotic members of the family (Bcl-2, Bcl-xL), allowing Bak and Bax to translocate to the outer mitochondrial membrane, thus permeabilizing it and facilitating release of pro-apoptotic proteins such as cytochrome c and Smac/DIABLO, an antagonist of inhibitors of apoptosis proteins (IAPs).
Soluble FasL is less active than its membrane-bound counterpart and does not induce receptor trimerization and DISC formation.
Functions
Apoptosis triggered by Fas-Fas ligand binding plays a fundamental role in the regulation of the immune system. Its functions include:
- T-cell homeostasis: the activation of T-cells leads to their expression of the Fas ligand. T cells are initially resistant to Fas-mediated apoptosis during clonal expansion, but become progressively more sensitive the longer they are activated, ultimately resulting in activation-induced cell death (AICD). This process is needed to prevent an excessive immune response and eliminate autoreactive T-cells. Humans and mice with deleterious mutations of Fas or Fas ligand develop an accumulation of aberrant T-cells, leading to lymphadenopathy, splenomegaly, and lupus erythematosus.
- Cytotoxic T-cell activity: Fas-induced apoptosis and the perforin pathway are the two main mechanisms by which cytotoxic T lymphocytes induce cell death in cells expressing foreign antigens.[4]
- Immune privilege: Cells in immune privileged areas such as the cornea or testes express Fas ligand and induce the apoptosis of infiltrating lymphocytes. It is one of many mechanisms the body employs in the establishment and maintenance of immune privilege.
- Maternal tolerance: Fas ligand may be instrumental in the prevention of leukocyte trafficking between the mother and the fetus, although no pregnancy defects have yet been attributed to a faulty Fas-Fas ligand system.
- Tumor counterattack: Tumors may over-express Fas ligand and induce the apoptosis of infiltrating lymphocytes, allowing the tumor to escape the effects of an immune response.[5] The up-regulation of Fas ligand often occurs following chemotherapy, from which the tumor cells have attained apoptosis resistance.
Role in disease
Defective Fas-mediated apoptosis may lead to oncogenesis as well as drug resistance in existing tumors. Germline mutation of Fas is associated with autoimmune lymphoproliferative syndrome (ALPS), a childhood disorder of apoptosis.
Interactions
Fas ligand has been shown to interact with:
See also
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Sheikh MS, Fornace AJ (2000). "Death and decoy receptors and p53-mediated apoptosis". Leukemia. 14 (8): 1509–1513. doi:10.1038/sj.leu.2401865. PMID 10942251.
- ↑ Andersen MH, Schrama D, Thor Straten P, Becker JC (2006). "Cytotoxic T cells". J. Invest. Dermatol. 126 (1): 32–41. doi:10.1038/sj.jid.5700001. PMID 16417215.
- ↑ Igney FH, Krammer PH (2005). "Tumor counterattack: fact or fiction?". Cancer Immunol. Immunother. 54 (11): 1127–1136. doi:10.1007/s00262-005-0680-7. PMID 15889255.
- 1 2 3 4 Gajate C, Mollinedo F (March 2005). "Cytoskeleton-mediated death receptor and ligand concentration in lipid rafts forms apoptosis-promoting clusters in cancer chemotherapy". J. Biol. Chem. 280 (12): 11641–7. doi:10.1074/jbc.M411781200. PMID 15659383.
- 1 2 3 Micheau O, Tschopp J (July 2003). "Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes". Cell. 114 (2): 181–90. doi:10.1016/s0092-8674(03)00521-x. PMID 12887920.
- ↑ Parlato S, Giammarioli AM, Logozzi M, Lozupone F, Matarrese P, Luciani F, Falchi M, Malorni W, Fais S (October 2000). "CD95 (APO-1/Fas) linkage to the actin cytoskeleton through ezrin in human T lymphocytes: a novel regulatory mechanism of the CD95 apoptotic pathway". EMBO J. 19 (19): 5123–34. doi:10.1093/emboj/19.19.5123. PMC 302100
 . PMID 11013215.
. PMID 11013215. - 1 2 3 Ghadimi MP, Sanzenbacher R, Thiede B, Wenzel J, Jing Q, Plomann M, Borkhardt A, Kabelitz D, Janssen O (May 2002). "Identification of interaction partners of the cytosolic polyproline region of CD95 ligand (CD178)". FEBS Lett. 519 (1-3): 50–8. doi:10.1016/s0014-5793(02)02709-6. PMID 12023017.
- 1 2 Wenzel J, Sanzenbacher R, Ghadimi M, Lewitzky M, Zhou Q, Kaplan DR, Kabelitz D, Feller SM, Janssen O (December 2001). "Multiple interactions of the cytosolic polyproline region of the CD95 ligand: hints for the reverse signal transduction capacity of a death factor". FEBS Lett. 509 (2): 255–62. doi:10.1016/s0014-5793(01)03174-x. PMID 11741599.
- ↑ Hane M, Lowin B, Peitsch M, Becker K, Tschopp J (October 1995). "Interaction of peptides derived from the Fas ligand with the Fyn-SH3 domain". FEBS Lett. 373 (3): 265–8. doi:10.1016/0014-5793(95)01051-f. PMID 7589480.
- ↑ Starling GC, Bajorath J, Emswiler J, Ledbetter JA, Aruffo A, Kiener PA (April 1997). "Identification of amino acid residues important for ligand binding to Fas". J. Exp. Med. 185 (8): 1487–92. doi:10.1084/jem.185.8.1487. PMC 2196280
 . PMID 9126929.
. PMID 9126929. - ↑ Schneider P, Bodmer JL, Holler N, Mattmann C, Scuderi P, Terskikh A, Peitsch MC, Tschopp J (July 1997). "Characterization of Fas (Apo-1, CD95)-Fas ligand interaction". J. Biol. Chem. 272 (30): 18827–33. doi:10.1074/jbc.272.30.18827. PMID 9228058.
- ↑ Yu KY, Kwon B, Ni J, Zhai Y, Ebner R, Kwon BS (May 1999). "A newly identified member of tumor necrosis factor receptor superfamily (TR6) suppresses LIGHT-mediated apoptosis". J. Biol. Chem. 274 (20): 13733–6. doi:10.1074/jbc.274.20.13733. PMID 10318773.
- ↑ Hsu TL, Chang YC, Chen SJ, Liu YJ, Chiu AW, Chio CC, Chen L, Hsieh SL (May 2002). "Modulation of dendritic cell differentiation and maturation by decoy receptor 3". J. Immunol. 168 (10): 4846–53. doi:10.4049/jimmunol.168.10.4846. PMID 11994433.
- ↑ Pitti RM, Marsters SA, Lawrence DA, Roy M, Kischkel FC, Dowd P, Huang A, Donahue CJ, Sherwood SW, Baldwin DT, Godowski PJ, Wood WI, Gurney AL, Hillan KJ, Cohen RL, Goddard AD, Botstein D, Ashkenazi A (December 1998). "Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer". Nature. 396 (6712): 699–703. doi:10.1038/25387. PMID 9872321.
Further reading
- Choi C, Benveniste EN (January 2004). "Fas ligand/Fas system in the brain: regulator of immune and apoptotic responses". Brain Res. Brain Res. Rev. 44 (1): 65–81. doi:10.1016/j.brainresrev.2003.08.007. PMID 14739003.
- Tolstrup M, Ostergaard L, Laursen AL, Pedersen SF, Duch M (2004). "HIV/SIV escape from immune surveillance: focus on Nef". Curr. HIV Res. 2 (2): 141–51. doi:10.2174/1570162043484924. PMID 15078178.
External links
- GeneReviews/NCBI/NIH/UW entry on Autoimmune Lymphoproliferative Syndrome
- Online Mendelian Inheritance in Man (OMIM) 601859
- Fas Ligand Protein at the US National Library of Medicine Medical Subject Headings (MeSH)