Programming with Big Data in R
| Paradigm | SPMD and MPMD |
|---|---|
| Designed by | Wei-Chen Chen, George Ostrouchov, Pragneshkumar Patel, and Drew Schmidt |
| Developer | pbdR Core Team |
| First appeared | Sep. 2012 |
| Preview release | |
| Typing discipline | Dynamic |
| OS | Cross-platform |
| License | General Public License and Mozilla Public License |
| Website |
www |
| Influenced by | |
| R, C, Fortran, MPI, and ØMQ | |
Programming with Big Data in R (pbdR)[1] is a series of R packages and an environment for statistical computing with Big Data by using high-performance statistical computation.[2] The pbdR uses the same programming language as R with S3/S4 classes and methods which is used among statisticians and data miners for developing statistical software. The significant difference between pbdR and R code is that pbdR mainly focuses on distributed memory systems, where data are distributed across several processors and analyzed in a batch mode, while communications between processors are based on MPI that is easily used in large high-performance computing (HPC) systems. R system mainly focuses on single multi-core machines for data analysis via an interactive mode such as GUI interface.
Two main implementations in R using MPI are Rmpi[3] and pbdMPI of pbdR.
- The pbdR built on pbdMPI uses SPMD parallelism where every processor is considered as worker and owns parts of data. The SPMD parallelism introduced in mid 1980 is particularly efficient in homogeneous computing environments for large data, for example, performing singular value decomposition on a large matrix, or performing clustering analysis on high-dimensional large data. On the other hand, there is no restriction to use manager/workers parallelism in SPMD parallelism environment.
- The Rmpi[3] uses manager/workers parallelism where one main processor (manager) servers as the control of all other processors (workers). The manager/workers parallelism introduced around early 2000 is particularly efficient for large tasks in small clusters, for example, bootstrap method and Monte Carlo simulation in applied statistics since i.i.d. assumption is commonly used in most statistical analysis. In particular, task pull parallelism has better performance for Rmpi in heterogeneous computing environments.
The idea of SPMD parallelism is to let every processor do the same amount of work, but on different parts of a large data set. For example, a modern GPU is a large collection of slower co-processors that can simply apply the same computation on different parts of relatively smaller data, but the SPMD parallelism ends up with an efficient way to obtain final solutions (i.e. time to solution is shorter).[4] It is clear that pbdR is not only suitable for small clusters, but is also more stable for analyzing Big data and more scalable for supercomputers.[5] In short, pbdR
- does not like Rmpi, snow, snowfall, do-like, nor parallel packages in R,
- does not focus on interactive computing nor master/workers,
- but is able to use both SPMD and task parallelisms.
Package design
Programming with pbdR requires usage of various packages developed by pbdR core team. Packages developed are the following.
| General | I/O | Computation | Application | Profiling | Client/Server |
|---|---|---|---|---|---|
| pbdDEMO | pbdNCDF4 | pbdDMAT | pmclust | pbdPROF | pbdZMQ |
| pbdMPI | pbdADIOS | pbdBASE | pbdML | pbdPAPI | remoter |
| pbdSLAP | hpcvis | pbdCS |
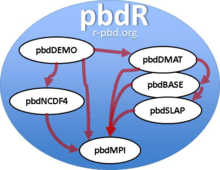
Among these packages, pbdMPI provides wrapper functions to MPI library, and it also produces a shared library and a configuration file for MPI environments. All other packages rely on this configuration for installation and library loading that avoids difficulty of library linking and compiling. All other packages can directly use MPI functions easily.
- pbdMPI --- an efficient interface to MPI either OpenMPI or MPICH2 with a focus on Single Program/Multiple Data (SPMD) parallel programming style
- pbdSLAP --- bundles scalable dense linear algebra libraries in double precision for R, based on ScaLAPACK version 2.0.2 which includes several scalable linear algebra packages (namely BLACS, PBLAS, and ScaLAPACK).
- pbdNCDF4 --- interface to Parallel Unidata NetCDF4 format data files
- pbdBASE --- low-level ScaLAPACK codes and wrappers
- pbdDMAT --- distributed matrix classes and computational methods, with a focus on linear algebra and statistics
- pbdDEMO --- set of package demonstrations and examples, and this unifying vignette
- pmclust --- parallel model-based clustering using pbdR
- pbdPROF --- profiling package for MPI codes and visualization of parsed stats
- pbdZMQ --- interface to ØMQ
Among those packages, the pbdDEMO package is a collection of 20+ package demos which offer example uses of the various pbdR packages, and contains a vignette that offers detailed explanations for the demos and provides some mathematical or statistical insight.
Examples
Example 1
Hello World! Save the following code in a file called "demo.r"
### Initial MPI
library(pbdMPI, quiet = TRUE)
init()
comm.cat("Hello World!\n")
### Finish
finalize()
and use the command
mpiexec -np 2 Rscript demo.r
to execute the code where Rscript is one of command line executable program.
Example 2
The following example modified from pbdMPI illustrates the basic syntax of the language of pbdR. Since pbdR is designed in SPMD, all the R scripts are stored in files and executed from the command line via mpiexec, mpirun, etc. Save the following code in a file called "demo.r"
### Initial MPI
library(pbdMPI, quiet = TRUE)
init()
.comm.size <- comm.size()
.comm.rank <- comm.rank()
### Set a vector x on all processors with different values
N <- 5
x <- (1:N) + N * .comm.rank
### All reduce x using summation operation
y <- allreduce(as.integer(x), op = "sum")
comm.print(y)
y <- allreduce(as.double(x), op = "sum")
comm.print(y)
### Finish
finalize()
and use the command
mpiexec -np 4 Rscript demo.r
to execute the code where Rscript is one of command line executable program.
Example 3
The following example modified from pbdDEMO illustrates the basic ddmatrix computation of pbdR which performs singular value decomposition on a given matrix. Save the following code in a file called "demo.r"
# Initialize process grid
library(pbdDMAT, quiet=T)
if(comm.size() != 2)
comm.stop("Exactly 2 processors are required for this demo.")
init.grid()
# Setup for the remainder
comm.set.seed(diff=TRUE)
M <- N <- 16
BL <- 2 # blocking --- passing single value BL assumes BLxBL blocking
dA <- ddmatrix("rnorm", nrow=M, ncol=N, mean=100, sd=10)
# LA SVD
svd1 <- La.svd(dA)
comm.print(svd1$d)
# Finish
finalize()
and use the command
mpiexec -np 2 Rscript demo.r
to execute the code where Rscript is one of command line executable program.
Further reading
- Raim, A.M. (2013). Introduction to distributed computing with pbdR at the UMBC High Performance Computing Facility (PDF) (Technical report). UMBC High Performance Computing Facility, University of Maryland, Baltimore County. HPCF-2013-2.
- Bachmann, M.G., Dyas, A.D., Kilmer, S.C. and Sass, J. (2013). Block Cyclic Distribution of Data in pbdR and its Effects on Computational Efficiency (PDF) (Technical report). UMBC High Performance Computing Facility, University of Maryland, Baltimore County. HPCF-2013-11.
- Bailey, W.J., Chambless, C.A., Cho, B.M. and Smith, J.D. (2013). Identifying Nonlinear Correlations in High Dimensional Data with Application to Protein Molecular Dynamics Simulations (PDF) (Technical report). UMBC High Performance Computing Facility, University of Maryland, Baltimore County. HPCF-2013-12.
- Dirk Eddelbuettel. "High-Performance and Parallel Computing with R".
- "R at 12,000 Cores".
This article was read 22,584 times in 2012 since it posted on October 16, 2012 and ranked number 3[6] - Google Summer of Code - R 2013. "Profiling Tools for Parallel Computing with R".
- Wush Wu (2014). "在雲端運算環境使用R和MPI".
- Wush Wu (2013). "快速在AWS建立R和pbdMPI的使用環境".
References
- ↑ Ostrouchov, G., Chen, W.-C., Schmidt, D., Patel, P. (2012). "Programming with Big Data in R".
- ↑ Chen, W.-C. & Ostrouchov, G. (2011). "HPSC -- High Performance Statistical Computing for Data Intensive Research".
- 1 2 Yu, H. (2002). "Rmpi: Parallel Statistical Computing in R". R News.
- ↑ Mike Houston. "Folding@Home - GPGPU". Retrieved 2007-10-04.
- ↑ Schmidt, D., Ostrouchov, G., Chen, W.-C., and Patel, P. (2012). "Tight Coupling of R and Distributed Linear Algebra for High-Level Programming with Big Data". High Performance Computing, Networking, Storage and Analysis (SCC), 2012 SC Companion:: 811–815.
- ↑ "100 most read R posts in 2012 (stats from R-bloggers) – big data, visualization, data manipulation, and other languages".