Hoogsteen base pair
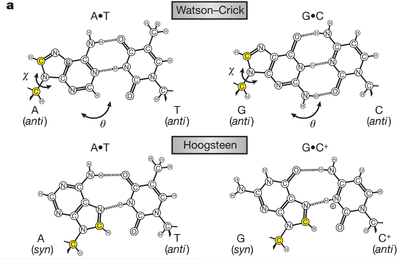
A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 position of the purine base (as a hydrogen bond acceptor) and C6 amino group (as a donor), which bind the Watson-Crick (N3–N4) face of the pyrimidine base.
History
Ten years after James Watson and Francis Crick published their model of the DNA double helix,[2] Karst Hoogsteen reported [3] a crystal structure of a complex in which analogues of A and T formed a base pair that had a different geometry from that described by Watson and Crick. Similarly, an alternative base-pairing geometry can occur for G•C pairs. Hoogsteen pointed out that if the alternative hydrogen-bonding patterns were present in DNA, then the double helix would have to assume a quite different shape. Hoogsteen base pairs are, however, rarely observed.
Chemical properties
Hoogsteen pairs have quite different properties from Watson-Crick base pairs. The angle between the two glycosidic bonds (ca. 80° in the A• T pair) is larger and the C1′–C1′ distance (ca. 860 pm or 8.6 Å) is smaller than in the regular geometry. In some cases, called reversed Hoogsteen base pairs, one base is rotated 180° with respect to the other. In some DNA sequences, especially CA and TA dinucleotides, Hoogsteen base pairs exist as transient entities that are present in thermal equilibrium with standard Watson–Crick base pairs. The detection of the transient species required the use of NMR techniques that have only recently been applied to macromolecules.[1]
Hoogsteen base pairs have been observed in protein–DNA complexes.[4] Some proteins have evolved to recognize only one base-pair type, and use intermolecular interactions to shift the equilibrium between the two geometries. DNA has many features that allow its sequence-specific recognition by proteins. This recognition was originally thought to primarily involve specific hydrogen-bonding interactions between amino-acid side chains and bases. But it soon became clear that there was no identifiable one-to-one correspondence — that is, there was no simple code to be read. Part of the problem is that DNA can undergo conformational changes that distort the classical double helix. The resulting variations in the way that DNA bases are presented to proteins can thus affect the recognition mechanism. It has become evident that distortions in the double helix are themselves dependent on base sequence. This enables proteins to recognize DNA shape in a manner reminiscent of the way that they recognize other proteins and small ligand molecules. For example, stretches of A and T bases can narrow the minor groove of DNA (the narrower of the two grooves in the double helix), thus enhancing local negative electrostatic potentials and creating binding sites for appropriately placed, positively charged arginine amino-acid residues.
Triplex structures
This non-Watson-Crick base-pairing allows the third strands to wind around the duplexes, which are assembled in the Watson-Crick pattern, and form triple-stranded helices such as (poly(dA)•2poly(dT)) and (poly(rG)•2poly(rC)). It can be also seen in three-dimensional structures of transfer RNA.
Quadruplex structures
It also allows formation of secondary structures of single stranded DNA and RNA G-rich called G-quadruplexes (G4-DNA and G4-RNA) at least in vitro. It needs four triplets of G, separated by short spacers. This permits assembly of planar quartets which are composed of stacked associations of hoogsteen bonded guanine molecules.[5]
Triple helix base pairing
Watson and Crick base pairs are indicated by a "•", "-", or a "." (example: A•T, or poly(rC)•2poly(rC)).
Hoogsteen triple helix base pairs are indicated by a "*" or a ":" (example: C•G*C+, T•A*T, C•G*G, or T•A*A).
See also
- Base pair
- Wobble base pair
- G-quadruplex
- RNA Tertiary Structure
- PPRHs, Polypurine Reverse Hoogsteen hairpins, oligonucleotides that can bind either DNA or RNA and decrease gene expression.
References
- 1 2 Evgenia N. Nikolova; Eunae Kim; Abigail A. Wise; Patrick J. O’Brien; Ioan Andricioaei; Hashim M. Al-Hashimi (2011). "Transient Hoogsteen base pairs in canonical duplex DNA". Nature. 470: 498–502. doi:10.1038/nature09775.
- ↑ Watson JD, Crick FH (1953). "Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid". Nature. 171 (4356): 737–738. doi:10.1038/171737a0. PMID 13054692.
- ↑ Hoogsteen K (1963). "The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine". Acta Crystallographica. 16: 907–916. doi:10.1107/S0365110X63002437.
- ↑ Jun Aishima, Rossitza K. Gitti, Joyce E. Noah, Hin Hark Gan, Tamar Schlick, Cynthia Wolberger (2002). "A Hoogsteen base pair embedded in undistorted B‐DNA". Nucl Acids Res. 30 (23): 5244–5252. doi:10.1093/nar/gkf661.
- ↑ Johnson JE, Smith JS, Kozak ML, Johnson FB (2008). "In vivo veritas: Using yeast to probe the biological functions of G-quadruplexes". Biochimie. 90 (8): 1250–1263. doi:10.1016/j.biochi.2008.02.013. PMC 2585026
 . PMID 18331848.
. PMID 18331848.