HMG-CoA reductase
| View/Edit Human | View/Edit Mouse |
| HMG-CoA reductase | |
|---|---|
| EC number | {{{EC_number}}} |
| Gene Ontology | AmiGO / EGO |
| hydroxymethylglutaryl-CoA reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.1.1.88 | ||||||||
| CAS number | 37250-24-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
| hydroxymethylglutaryl-CoA reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.1.1.34 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
HMG-CoA reductase (3-hydroxy-3-methyl-glutaryl-coenzyme A reductase, officially abbreviated HMGCR) is the rate-controlling enzyme (NADH-dependent, EC 1.1.1.88; NADPH-dependent, EC 1.1.1.34) of the mevalonate pathway, the metabolic pathway that produces cholesterol and other isoprenoids. Normally in mammalian cells this enzyme is suppressed by cholesterol derived from the internalization and degradation of low density lipoprotein (LDL) via the LDL receptor as well as oxidized species of cholesterol. Competitive inhibitors of the reductase induce the expression of LDL receptors in the liver, which in turn increases the catabolism of plasma LDL and lowers the plasma concentration of cholesterol, an important determinant of atherosclerosis.[4] This enzyme is thus the target of the widely available cholesterol-lowering drugs known collectively as the statins.
HMG-CoA reductase is anchored in the membrane of the endoplasmic reticulum, and was long regarded as having seven transmembrane domains, with the active site located in a long carboxyl terminal domain in the cytosol. More recent evidence shows it to contain eight transmembrane domains.[5]
In humans, the gene for HMG-CoA reductase is located on the long arm of the fifth chromosome (5q13.3-14).[6] Related enzymes having the same function are also present in other animals, plants and bacteria.
Structure
The main isoform (isoform 1) of HMG-CoA reductase in humans is 888 amino acids long. It is a polytopic transmembrane protein (meaning it possesses many alpha helical transmembrane segments). It contains two main domains:
- an N-terminal sterol-sensing domain (amino acid interval: 88-218), which binds sterol groups. Cholesterol binding at this region inhibits the activity of the catalytic domain.
- a C-terminal catalytic domain (amino acid interval: 489-871), namely the 3-hydroxy-3-methyl-glutaryl-CoA reductase domain. This domain is required for the proper enzymatic activity of the protein.
Isoform 2 is 835 amino acids long. This variant is shorter because it lacks an exon in the middle region. This does not affect any of the aforementioned domains.
Function
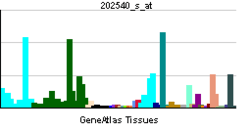
HMGCR catalyses the conversion of HMG-CoA to mevalonic acid, a necessary step in the biosynthesis of cholesterol.:
 |
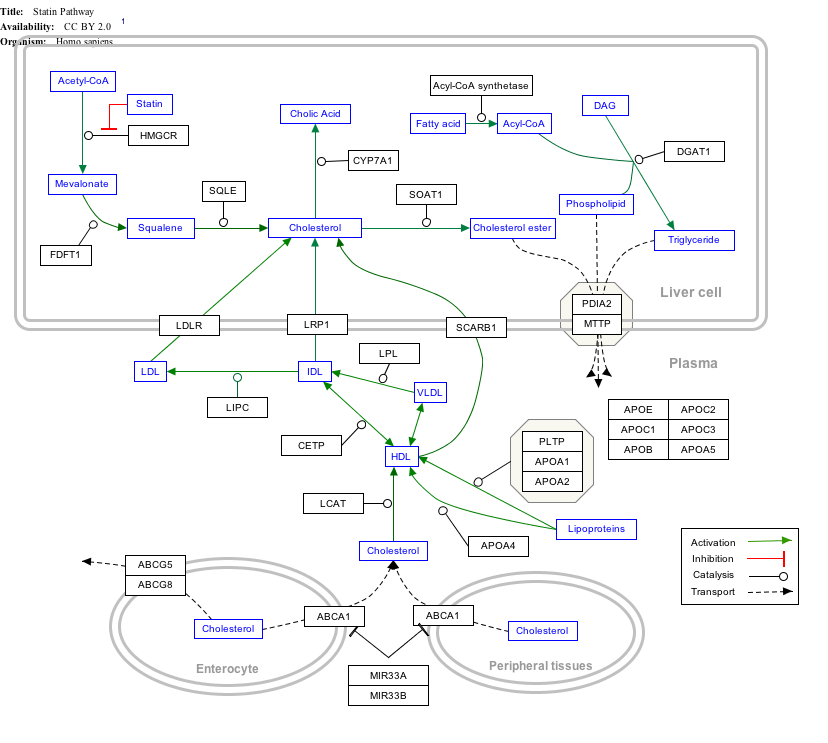
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
Statin Pathway edit
- ↑ The interactive pathway map can be edited at WikiPathways: "Statin_Pathway_WP430".
Inhibitors
Drugs
Drugs that inhibit HMG-CoA reductase, known collectively as HMG-CoA reductase inhibitors (or "statins"), are used to lower serum cholesterol as a means of reducing the risk for cardiovascular disease.[7]
These drugs include rosuvastatin (CRESTOR), lovastatin (Mevacor), atorvastatin (Lipitor), pravastatin (Pravachol), fluvastatin (Lescol), pitavastatin (Livalo), and simvastatin (Zocor).[8] Red yeast rice extract, one of the fungal sources from which the statins were discovered, contains several naturally occurring cholesterol-lowering molecules known as monacolins. The most active of these is monacolin K, or lovastatin (previously sold under the trade name Mevacor, and now available as generic lovastatin).[9]
Vytorin is drug that combines the use simvastatin and ezetimibe, which slows the formation of cholesterol by every cell in the body, along with ezetimibe reducing absorption of cholesterol, typically by about 53%, from the intestines.[10]
Hormones
HMG-CoA reductase is active when blood glucose is high. The basic functions of insulin and glucagon are to maintain glucose homeostasis. Thus, in controlling blood sugar levels, they indirectly affect the activity of HMG-CoA reductase, but a decrease in activity of the enzyme is caused by an AMP-activated protein kinase, which responds to an increase in AMP concentration, and also to leptin (see 4.4, Phosphorylation of reductase).
Clinical significance
Since the reaction catalysed by HMG-CoA reductase is the rate-limiting step in cholesterol synthesis, this enzyme represents the sole major drug target for contemporary cholesterol-lowering drugs in humans. The medical significance of HMG-CoA reductase has continued to expand beyond its direct role in cholesterol synthesis following the discovery that statins can offer cardiovascular health benefits independent of cholesterol reduction.[11] Statins have been shown to have anti-inflammatory properties,[12] most likely as a result of their ability to limit production of key downstream isoprenoids that are required for portions of the inflammatory response. It can be noted that blocking of isoprenoid synthesis by statins has shown promise in treating a mouse model of multiple sclerosis, an inflammatory autoimmune disease.[13]
HMG-CoA reductase is an important developmental enzyme. Inhibition of its activity and the concomitant lack of isoprenoids that yields can lead to germ cell migration defects [14] as well as intracerebral hemorrhage.[15]
Regulation

Regulation of HMG-CoA reductase is achieved at several levels: transcription, translation, degradation and phosphorylation.
Transcription of the reductase gene
Transcription of the reductase gene is enhanced by the sterol regulatory element binding protein (SREBP). This protein binds to the sterol regulatory element (SRE), located on the 5' end of the reductase gene. When SREBP is inactive, it is bound to the ER or nuclear membrane with another protein called SREBP cleavage-activating protein (SCAP). When cholesterol levels fall, SREBP is released from the membrane by proteolysis and migrates to the nucleus, where it binds to the SRE and transcription is enhanced. If cholesterol levels rise, proteolytic cleavage of SREBP from the membrane ceases and any proteins in the nucleus are quickly degraded.
Translation of mRNA
Translation of mRNA is inhibited by a mevalonate derivative, which has been reported to be farnesol,[16][17] although this role has been disputed.[18]
Degradation of reductase
Rising levels of sterols increase the susceptibility of the reductase enzyme to ER-associated degradation (ERAD) and proteolysis. Helices 2-6 (total of 8) of the HMG-CoA reductase transmembrane domain sense the higher levels of cholesterol, which leads to the exposure of Lysine 248. This lysine residue can become ubiquinated by the E3 ligase AMFR, serving as a signal for proteolytic degradation.
Phosphorylation of reductase
Short-term regulation of HMG-CoA reductase is achieved by inhibition by phosphorylation (of Serine 872, in humans[19]). Decades ago it was believed that a cascade of enzymes controls the activity of HMG-CoA reductase: an HMG-CoA reductase kinase was thought to inactivate the enzyme, and the kinase in turn was held to be activated via phosphorylation by HMG-CoA reductase kinase kinase. An excellent review on regulation of the mevalonate pathway by Nobel Laureates Joseph Goldstein and Michael Brown adds specifics: HMG-CoA reductase is phosphorylated and inactivated by an AMP-activated protein kinase, which also phosphorylates and inactivates acetyl-CoA carboxylase, the rate-limiting enzyme of fatty acid biosynthesis.[20] Thus, both pathways utilizing acetyl-CoA for lipid synthesis are inactivated when energy charge is low in the cell, and concentrations of AMP rise. There has been a great deal of research on the identity of upstream kinases that phosphorylate and activate the AMP-activated protein kinase.[21]
Fairly recently, LKB1 has been identified as a likely AMP kinase kinase,[22] which appears to involve calcium/calmodulin signaling. This pathway likely transduces signals from leptin, adiponectin, and other signaling molecules.[21]
See also
References
- ↑ "Drugs that physically interact with 3-hydroxy-3-methylglutaryl-coenzyme A reductase view/edit references on wikidata".
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ "Entrez Gene: HMGCR 3-hydroxy-3-methylglutaryl-Coenzyme A reductase".
- ↑ Roitelman J, Olender EH, Bar-Nun S, Dunn WA, Simoni RD (Jun 1992). "Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum". The Journal of Cell Biology. 117 (5): 959–73. doi:10.1083/jcb.117.5.959. PMC 2289486
 . PMID 1374417.
. PMID 1374417. - ↑ Lindgren V, Luskey KL, Russell DW, Francke U (Dec 1985). "Human genes involved in cholesterol metabolism: chromosomal mapping of the loci for the low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl-coenzyme A reductase with cDNA probes". Proceedings of the National Academy of Sciences of the United States of America. 82 (24): 8567–71. doi:10.1073/pnas.82.24.8567. PMC 390958
 . PMID 3866240.
. PMID 3866240. - ↑ Farmer JA (1998). "Aggressive lipid therapy in the statin era". Progress in Cardiovascular Diseases. 41 (2): 71–94. doi:10.1016/S0033-0620(98)80006-6. PMID 9790411.
- ↑ "Is there a "best" statin drug?". The Johns Hopkins Medical Letter Health After 50. 15 (11): 4–5. Jan 2004. PMID 14983817.
- ↑ Lin YL, Wang TH, Lee MH, Su NW (Jan 2008). "Biologically active components and nutraceuticals in the Monascus-fermented rice: a review". Applied Microbiology and Biotechnology. 77 (5): 965–73. doi:10.1007/s00253-007-1256-6. PMID 18038131.
- ↑ Flores NA (Sep 2004). "Ezetimibe + simvastatin (Merck/Schering-Plough)". Current Opinion in Investigational Drugs. 5 (9): 984–92. PMID 15503655.
- ↑ Arnaud C, Veillard NR, Mach F (Apr 2005). "Cholesterol-independent effects of statins in inflammation, immunomodulation and atherosclerosis". Current Drug Targets. Cardiovascular & Haematological Disorders. 5 (2): 127–34. doi:10.2174/1568006043586198. PMID 15853754.
- ↑ Sorrentino S, Landmesser U (Dec 2005). "Nonlipid-lowering effects of statins". Current Treatment Options in Cardiovascular Medicine. 7 (6): 459–466. doi:10.1007/s11936-005-0031-1. PMID 16283973.
- ↑ Stüve O, Youssef S, Steinman L, Zamvil SS (Jun 2003). "Statins as potential therapeutic agents in neuroinflammatory disorders". Current Opinion in Neurology. 16 (3): 393–401. doi:10.1097/01.wco.0000073942.19076.d1 (inactive 2015-01-01). PMID 12858078.
- ↑ Thorpe JL, Doitsidou M, Ho SY, Raz E, Farber SA (Feb 2004). "Germ cell migration in zebrafish is dependent on HMGCoA reductase activity and prenylation". Developmental Cell. 6 (2): 295–302. doi:10.1016/S1534-5807(04)00032-2. PMID 14960282.
- ↑ Eisa-Beygi S, Hatch G, Noble S, Ekker M, Moon TW (Jan 2013). "The 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) pathway regulates developmental cerebral-vascular stability via prenylation-dependent signalling pathway". Developmental Biology. 373 (2): 258–266. doi:10.1016/j.ydbio.2012.11.024. PMID 23206891.
- ↑ Meigs TE, Roseman DS, Simoni RD (Apr 1996). "Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation by the nonsterol mevalonate metabolite farnesol in vivo". The Journal of Biological Chemistry. 271 (14): 7916–22. doi:10.1074/jbc.271.14.7916. PMID 8626470.
- ↑ Meigs TE, Simoni RD (Sep 1997). "Farnesol as a regulator of HMG-CoA reductase degradation: characterization and role of farnesyl pyrophosphatase". Archives of Biochemistry and Biophysics. 345 (1): 1–9. doi:10.1006/abbi.1997.0200. PMID 9281305.
- ↑ Keller RK, Zhao Z, Chambers C, Ness GC (Apr 1996). "Farnesol is not the nonsterol regulator mediating degradation of HMG-CoA reductase in rat liver". Archives of Biochemistry and Biophysics. 328 (2): 324–30. doi:10.1006/abbi.1996.0180. PMID 8645011.
- ↑ Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J (Mar 2000). "Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis". The EMBO Journal. 19 (5): 819–30. doi:10.1093/emboj/19.5.819. PMC 305622
 . PMID 10698924.
. PMID 10698924. - ↑ Goldstein JL, Brown MS (Feb 1990). "Regulation of the mevalonate pathway". Nature. 343 (6257): 425–30. doi:10.1038/343425a0. PMID 1967820.
- 1 2 Hardie DG, Scott JW, Pan DA, Hudson ER (Jul 2003). "Management of cellular energy by the AMP-activated protein kinase system". FEBS Letters. 546 (1): 113–20. doi:10.1016/S0014-5793(03)00560-X. PMID 12829246.
- ↑ Witters LA, Kemp BE, Means AR (Jan 2006). "Chutes and Ladders: the search for protein kinases that act on AMPK". Trends in Biochemical Sciences. 31 (1): 13–6. doi:10.1016/j.tibs.2005.11.009. PMID 16356723.
Further reading
- Hodge VJ, Gould SJ, Subramani S, Moser HW, Krisans SK (Dec 1991). "Normal cholesterol synthesis in human cells requires functional peroxisomes". Biochemical and Biophysical Research Communications. 181 (2): 537–41. doi:10.1016/0006-291X(91)91222-X. PMID 1755834.
- Ramharack R, Tam SP, Deeley RG (Nov 1990). "Characterization of three distinct size classes of human 3-hydroxy-3-methylglutaryl coenzyme A reductase mRNA: expression of the transcripts in hepatic and nonhepatic cells". DNA and Cell Biology. 9 (9): 677–90. doi:10.1089/dna.1990.9.677. PMID 1979742.
- Clarke PR, Hardie DG (Aug 1990). "Regulation of HMG-CoA reductase: identification of the site phosphorylated by the AMP-activated protein kinase in vitro and in intact rat liver". The EMBO Journal. 9 (8): 2439–46. PMC 552270
 . PMID 2369897.
. PMID 2369897. - Luskey KL, Stevens B (Aug 1985). "Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation". The Journal of Biological Chemistry. 260 (18): 10271–7. PMID 2991281.
- Humphries SE, Tata F, Henry I, Barichard F, Holm M, Junien C, Williamson R (1986). "The isolation, characterisation, and chromosomal assignment of the gene for human 3-hydroxy-3-methylglutaryl coenzyme A reductase, (HMG-CoA reductase)". Human Genetics. 71 (3): 254–8. doi:10.1007/BF00284585. PMID 2998972.
- Beg ZH, Stonik JA, Brewer HB (Sep 1987). "Phosphorylation and modulation of the enzymic activity of native and protease-cleaved purified hepatic 3-hydroxy-3-methylglutaryl-coenzyme A reductase by a calcium/calmodulin-dependent protein kinase". The Journal of Biological Chemistry. 262 (27): 13228–40. PMID 3308873.
- Osborne TF, Goldstein JL, Brown MS (Aug 1985). "5' end of HMG CoA reductase gene contains sequences responsible for cholesterol-mediated inhibition of transcription". Cell. 42 (1): 203–12. doi:10.1016/S0092-8674(85)80116-1. PMID 3860301.
- Lindgren V, Luskey KL, Russell DW, Francke U (Dec 1985). "Human genes involved in cholesterol metabolism: chromosomal mapping of the loci for the low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl-coenzyme A reductase with cDNA probes". Proceedings of the National Academy of Sciences of the United States of America. 82 (24): 8567–71. doi:10.1073/pnas.82.24.8567. PMC 390958
 . PMID 3866240.
. PMID 3866240. - Lehoux JG, Kandalaft N, Belisle S, Bellabarba D (Oct 1985). "Characterization of 3-hydroxy-3-methylglutaryl coenzyme A reductase in human adrenal cortex". Endocrinology. 117 (4): 1462–8. doi:10.1210/endo-117-4-1462. PMID 3896758.
- Boguslawski W, Sokolowski W (1984). "HMG-CoA reductase activity in the microsomal fraction from human placenta in early and term pregnancy". The International Journal of Biochemistry. 16 (9): 1023–6. doi:10.1016/0020-711X(84)90120-4. PMID 6479432.
- Harwood HJ, Schneider M, Stacpoole PW (Sep 1984). "Measurement of human leukocyte microsomal HMG-CoA reductase activity". Journal of Lipid Research. 25 (9): 967–78. PMID 6491541.
- Nguyen LB, Salen G, Shefer S, Bullock J, Chen T, Tint GS, Chowdhary IR, Lerner S (Jul 1994). "Deficient ileal 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in sitosterolemia: sitosterol is not a feedback inhibitor of intestinal cholesterol biosynthesis". Metabolism. 43 (7): 855–9. doi:10.1016/0026-0495(94)90266-6. PMID 8028508.
- Bennis F, Favre G, Le Gaillard F, Soula G (Oct 1993). "Importance of mevalonate-derived products in the control of HMG-CoA reductase activity and growth of human lung adenocarcinoma cell line A549". International Journal of Cancer. Journal International Du Cancer. 55 (4): 640–5. doi:10.1002/ijc.2910550421. PMID 8406993.
- Van Doren M, Broihier HT, Moore LA, Lehmann R (Dec 1998). "HMG-CoA reductase guides migrating primordial germ cells". Nature. 396 (6710): 466–9. doi:10.1038/24871. PMID 9853754.
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES (Jul 1999). "Characterization of single-nucleotide polymorphisms in coding regions of human genes". Nature Genetics. 22 (3): 231–8. doi:10.1038/10290. PMID 10391209.
- Aboushadi N, Engfelt WH, Paton VG, Krisans SK (Sep 1999). "Role of peroxisomes in isoprenoid biosynthesis". The Journal of Histochemistry and Cytochemistry. 47 (9): 1127–32. doi:10.1177/002215549904700904. PMID 10449533.
- Honda A, Salen G, Honda M, Batta AK, Tint GS, Xu G, Chen TS, Tanaka N, Shefer S (Feb 2000). "3-Hydroxy-3-methylglutaryl-coenzyme A reductase activity is inhibited by cholesterol and up-regulated by sitosterol in sitosterolemic fibroblasts". The Journal of Laboratory and Clinical Medicine. 135 (2): 174–9. doi:10.1067/mlc.2000.104459. PMID 10695663.
- Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J (Mar 2000). "Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis". The EMBO Journal. 19 (5): 819–30. doi:10.1093/emboj/19.5.819. PMC 305622
 . PMID 10698924.
. PMID 10698924. - Istvan ES, Deisenhofer J (May 2001). "Structural mechanism for statin inhibition of HMG-CoA reductase". Science. 292 (5519): 1160–4. doi:10.1126/science.1059344. PMID 11349148.
- Rasmussen LM, Hansen PR, Nabipour MT, Olesen P, Kristiansen MT, Ledet T (Dec 2001). "Diverse effects of inhibition of 3-hydroxy-3-methylglutaryl-CoA reductase on the expression of VCAM-1 and E-selectin in endothelial cells". The Biochemical Journal. 360 (Pt 2): 363–70. doi:10.1042/0264-6021:3600363. PMC 1222236
 . PMID 11716764.
. PMID 11716764.
External links
- Cholesterol Synthesis - has some good regulatory details
- Proteopedia HMG-CoA_Reductase - the HMG-CoA Reductase Structure in Interactive 3D