Cell-free fetal DNA
Cell-free fetal DNA (cffDNA) is fetal DNA circulating freely in the maternal blood stream. It can be sampled by venipuncture on the mother. Analysis of cffDNA provides a method of non-invasive prenatal diagnosis and testing.
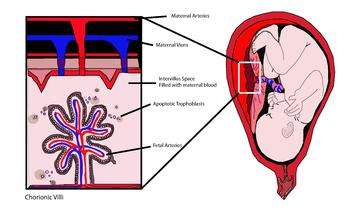
cffDNA originates from the trophoblasts making up the placenta.[1][2] An average of 11%-13.4% of cell-free DNA in maternal blood is of fetal origin, although this varies widely amongst patients.[3] The fetal DNA is fragmented and makes its way into the maternal bloodstream via shedding of the placental microparticles into the maternal bloodstream (figure 1).[4] Studies have shown that cffDNA can first be observed as early as 7 weeks gestation, and the amount of cffDNA increases as the pregnancy progresses.[5] cffDNA diminishes quickly after the birth of the baby, so that it is no longer detectable in the maternal blood approximately 2 hours after birth.[6] cffDNA is significantly smaller than the maternal DNA in the bloodstream, with fragments approximately 200bp in size.[7] Many protocols to extract the fetal DNA from the maternal plasma use its size to distinguish it from the maternal DNA.[8][9]
Studies have looked at, and some even optimized, protocols for testing non-compatible RhD factors, sex determination for X-linked genetic disorders and testing for single gene disorders.[10] Current studies are now looking at determining aneuploidies in the developing fetus.[11][12] These protocols can be done earlier than the current prenatal testing methods, and have no risk of spontaneous abortion, unlike current prenatal testing methods.[13] Non-invasive prenatal diagnosis (NIPD) and Non-invasive prenatal testing (NIPT) have been implemented in the UK and parts of the US; it has clear benefits above the standard tests of chorionic villi sample (CVS), amniocentesis,[14] and other diseases which have procedure-related miscarriage risks of about 1 in 100 pregnancies and 1 in 200 pregnancies, respectively.[15][16][17]
As a method of prenatal diagnosis, cell-free fetal DNA techniques share the same ethical and practical issues, such as the possibility of prenatal sex discernment and sex selection.[10]
Methods: From Extraction to Diagnosis
The general procedure is that a sample of the woman’s blood is taken after 10 weeks of pregnancy, although fetal blood can be detected as early as the fifth week.[18]
Limitations include the concentration of all cell-free DNA in maternal blood is low, the total amount of cell-free DNA varies between individuals, cell free fetal DNA molecules are out-numbered by maternal cell-free DNA molecules, the fetus inherits half the genome from the mother. However, there are ways around these limitations.[19]
Cell-free DNA purification
Protocols for isolation of plasma from maternal blood include centrifugation, followed by isolation and purification of cell-free DNA.[20] A standardized protocol by Legler et al. was created in 2007 from evaluation of previous primary literature. The highest yield for extraction of cell-free DNA was obtained with QIAamp DSP Virus Kit.[21]
Addition of formaldehyde to maternal blood samples increases the percentage of free fetal DNA. The purpose of formaldehyde is to stabilize intact cells, and inhibit further release of maternal DNA. The mean percentage of free fetal DNA in maternal bood ranges from 0.32% to 40%, with a mean percentage of 7.7[22] without formaldehyde-treatment. The mean percentage of free fetal DNA with formaldehyde treatment was 20.2%. However, Benachi et al. in 2005 and Chinnapapagari et al. 2005 have highlighted the range of results from 5% to 96%.[23][24] Another way to increase the fetal DNA is based on physical length of DNA fragments. Fetal DNA is smaller in size, a standardized size fractionation can be[8] and can comprise up to 70% of total cell-free DNA.
Specification of fetal DNA and mutation detection
To detect fetal DNA, the majority of studies focus on detecting paternally inherited sequences.[25][26][27] For example, primers can be designed to target the Y chromosome of male fetuses for polymerase chain reaction (PCR). A variety of methods have been used for mutation detection in fetal DNA.
Real-time quantitative PCR
Fluorescent probes are used to monitor the accumulation of amplicons produced throughout the PCR process. Thus, increase in reporter fluorescent signal is proportional to the number of amplicons generated. The appropriate real-time PCR protocol is designed according to the mutation or genotype to be detected. Point mutations are analysed with qualitative real-time PCR with the use of allele-specific probes. Insertions and deletions are analyzed by dosage measurements using quantitative real-time PCR.
Performance of RT-qPCR was examined by Hill and colleagues in 2010.[28] They analyzed SRY or DYS14 gene in cffDNA in 511 pregnancies. There was a high concordance rate of 401/403 for pregnancies at 7 weeks gestation or greater. Real-time PCR assays for single cell analysis have been developed for a Y-chromosome marker;[29] a common Tay-Sachs disease mutation,[30] the most common cystic fibrosis mutation[31] and a wide range of thalassemia mutations and Hb S.[32]
Nested PCR
Al-Yatama et al. 2010 evaluated the use of nested PCR to detect the Y chromosome in fetal DNA of maternal plasma. They detected 53/55 male fetuses. 3/25 women with female fetuses had a Y chromosome-specific signal. Thus, the results show 96% sensitivity and 88% specificity.[26]
Digital PCR
Microfluidic devices have the ability to perform highly parallel analysis in a single PCR step.[33] Absolute quantification can be achieved rather than relative quantification compared to RT-PCR. Thus, point mutations, copy number variations, loss of heterozygosity or aneuploidy can be detected.[12][34] Digital PCR can differentiate between maternal blood plasma and fetal DNA in a multiplex fashion.[33]
Shotgun sequencing
High-throughput shotgun sequencing technology from plasma of pregnant women, obtaining about 5 million sequence tags per patient sample can be done. Fan et al. 2008, using this method, were able to identify aneuploid pregnancies; trisomy detected at gestational ages as early as 14th week. Shotgun sequencing can be done with a Solexa/Illumina platform. Whole fetal genome mapping by parental haplotype analysis using sequencing of cell free fetal DNA was done in 2010.[16] Chiu et al. in 2010 studied 753 pregnant females, using a 2-plex massively parallel maternal plasma DNA sequencing and trisomy was diagnosed with z-score greater than 3.[35] The test was 100% sensitivity, 97.9% specificity, positive predictive value of 96.6%, and negative predictive value of 100%.
Mass spectrometry
Matrix-assisted laser desorption ionization/time of flight mass spectrometry (MALDI-TOF MS) combined with base extension after PCR allows cell free fetal DNA detection with single base specificity and single DNA molecule sensitivity.[36] DNA is first amplified by PCR, then linear amplification with base extension reaction (with a third primer) is designed to anneal to the region upstream of the mutation site. Either 1-2 bases are added to the extension primer to produce two extension products from wild-type DNA and mutant DNA. Since single base specificity is achieved, it is better than hybridization-based techniques with Taqman probes. According to Ding et al., no false negatives were found and no false positives were detected when looking for fetal DNA across 16 individuals.[36] A study by Akolekar and colleagues in 2010 genders of 90/91 males babies were confirmed, MALDI-TOF mass spectrometry had a 99.1% accuracy with 98.9% sensitivity, 99.2% specificity.[37]
Epigenetic modifications
Differences in gene activation between maternal DNA and fetal DNA can be exploited. Most current research is focussed on using epigenetic modifications to detect cffDNA.[38][39] Hypermethylated RASSF1A promoter has been reported as universal fetal marker to confirm the presence of cffDNA.[40] A new method was proposed by White et al. (2012). cffDNA was extracted from the maternal plasma, digested with methylation-sensitive and insensitive restriction enzymes and RT-PCR analysis of RASSF1A, SRY, DYS14 was done.[40] 79/90 (88%) of maternal blood samples were detected for hypermethylated RASSF1A.
Detection of mRNA derived from genes
mRNA transcripts from genes expressed in the placenta is detectable in maternal plasma.[41] The mixture of plasma is centrifuged and aqueous layer transferred, RNA is extracted. RT-PCR is set up for selected RNA expression. Specifically, hPL and beta-hCG mRNA is stable in maternal blood (Ng et al. 2002). This can help to confirm the presence of fetal DNA in the maternal plasma.[19]
Current use of cffdna
Paternity tests
Noninvasive prenatal paternity tests are available, for example from Natera and other companies. These tests can be done as soon as nine weeks after conception.
Early Identification of Fetal Sex
The identification of fetal sex for women who are carriers of X-linked diseases is advantageous for the family. X-linked diseases occur 5 in 10,000 live births.[42] The most commonly published sex-linked diseases include Duchenne muscular dystrophy and hemophilia. Ultrasonography is unreliable during the first trimester of pregnancy, as the genitalia are not fully developed. Other methods for sex determination before the use of cffDNA are invasive and performed at 11 weeks of gestation. There is a small risk of miscarriage.[43] The main method is to target the SRY gene on the Y chromosome and DYS14 sequence.[44][45]
Most X-linked diseases are evident in males because they lack the second X-chromosome that can compensate for the diseases allele. Some X-linked diseases include fragile-X syndrome, Duchenne muscular dystrophy and Hemophilia. In the case of X-linked diseases, if cffDNA can determine gender, invasive testing can be eliminated.[44] In congenital adrenal hyperplasia, the adrenal cortex lacks appropriate corticosteroid synthesis, leading to excess adrenal androgens and affects female fetuses.[46] There is an external masculinization of the genitalia in the female fetuses.[47] At the moment, at-risk mothers are given dexamethasone at 6 weeks of the pregnancy which suppresses the pituitary to release androgens. Invasive testing is used at 11 weeks of the pregnancy to determine the sex of the fetus. In Europe, tests for sex determination are available as early as 7 weeks of gestation; these tests cost $390.[48]
- Lack of the Y chromosome in the maternal plasma suggests that the fetus is female, but could also indicate failure to detect cffDNA in the maternal plasma. Thus, paternal polymorphisms or sex independent markers are further used to detect cffDNA. High heterozygosity of these markers must be detected for them to be application.[49]
Genetic studies for families at high risk for inherited genetic disorders
Severe monogenic diseases for which prenatal diagnosis is more commonly applied to include cystic fibrosis, beta-thalassemia, sickle cell anemia, spinal muscular atrophy, myotonic dystrophy, fragile-X syndrome, Duchenne muscular dystrophy and Hemophilia.[25] Both autosomal dominant and recessive disorders have been detected noninvasively by analyzing paternally inherited DNA.[44] Limitations to single gene disorders are autosomal recessive mutation or when the fetal autosomal dominant mutation is maternally inherited. There are also large sequence mutations that include duplication, expansion, insertion of DNA sequences.[50] cffDNA is fragmented with 200-300bp in length, thus these are harder to detect. One example is achondroplasia, a common autosomal dominant form of dwarfism, caused by FGFR3 gene point mutations.[51] Two pregnancies were examined in this study, one fetus was found to have the paternally inherited G1138A mutation and the other with a G1138A de novo mutation.[51] Another example is the Huntington's disease. It is currently diagnosed at 10–13 weeks at gestation with chronic villus sampling for polymorphic repeats. With qRT-PCR, there has been detection of CAG repeats at 17, 20 and 24, all normal levels.[52]
Routine prenatal screening for Rhesus factor
Mixing of fetal cells carrying paternal RhD antigens into maternal blood may result in the sensitization of an RhD-negative mother. Rhesus blood group (D antigen) is used to determine the risk of hemolytic disease in the fetus. In hemolytic disease, the maternal antibodies destroy RhD-positive fetal red blood cells. This leads to lethality for the fetus. A significant amount of blood can be exchanged between mother and infant during birth, CVS, amniocentesis and accidents.[19] 50 defined antigens on the surface of red blood cells indicate Rhesus blood group and E antigens are one of the most important. RHD gene determines the Rhesus D status.[53] Chinen et al. 2010 shows that 15% of Caucasian females, 3-5% of black African females and <3% of Asian females are RhD-negative. In the United Kingdom and other countries, cffDNA tests are now routinely being offered to RhD-negative patients at increased risk of isoimmunization. Anti-RhD immune globulin, a blood derivative, is only offered in the event of a RhD-positive fetus for these women (21, 48, 54).[19][46][54] In United States, prophylactic treatment is recommended for all RhD-negative pregnant women to prevent isoimmunization in case of RhD incompatibility.[55] In the United States, amniocentesis still serves as the gold standard diagnostic tool for those women who require antenatal fetal blood genotyping, but it has been suggested that technology using cell-free fetal DNA may ultimately replace this invasive procedure.[55]
Routine prenatal screening for aneuploidy
Aneuploidy, which refers to abnormal number of chromosomes, can also be detected using non-invasive prenatal tests. Research previous shown an increase in quantity of cffDNA in maternal plasma for fetal trisomy 13 and trisomy 21,[56] and it is not elevated in fetal trisomy 18.[57] A number of fetal nucleic acid molecules derived from aneuploid chromosomes can be detected including SERPINEB2 mRNA, clad B, hypomethylated SERPINB5 from chromosome 18, placenta-specific 4 (PLAC4), hypermethylated holocarboxylase synthetase (HLCS) and c21orf105 mRNA from chromosome 12.[58] With complete trisomy, the mRNA alleles in maternal plasma isn't the normal 1:1 ratio, but is in fact 2:1. Allelic ratios determined by epigenetic markers can also be used to detect the complete trisomies. Massive parallel sequencing and digital PCR for fetal aneuploidy detection can be used without restriction to fetal-specific nucleic acid molecules. Several cell-free fetal DNA and RNA technologies are under development to test a pregnancy for aneuploidy, mostly focusing on Down syndrome testing. Sampling of cffDNA from maternal blood for analysis by massively parallel sequencing (MPSS) is estimated to have a sensitivity of between 96 and 100%, and a specificity between 94 and 100% for detecting Down syndrome.[59] It can be performed at 10 weeks of gestational age.[60] One study in the United States estimated a false positive rate of 0.3% and a positive predictive value of 80% when using cffDNA to detect Down syndrome.[61]
Identification of Preeclampsia
Preeclampsia refers to abnormal placental growth which can cause multiple problems including preterm labor, polyhydramnios, hyperemesis gravidarum, and fetal-maternal hemorrhage.[19] Preeclampsia is the most prevalent pregnancy complication and predominant cause of premature birth, occurring at 2-5% of pregnancies in first world countries and 10% in developing countries.[19]
Lo et al. in 1999 studied quantitative variations of cffDNA in maternal blood for preeclampsia.[62] cffDNA was measured by using PCR to the SRY gene. The levels were fivefold more in preeclampsia pregnancies than normal. This is consistent with subsequent studies.[63][64] The largest study compared 120 women with preeclampsia pregnancies and 120 gestational age matched normal women. The cffDNA concentrations were 176 vs 75 genome equivalents/mL at 29 weeks of gestations. cffDNA concentration can be used as a screening tool.
Future perspectives
Cell-free fetal DNA can be used for whole genome sequencing, thus determining the complete DNA sequence of every gene of the baby.[65] This will be more clinically useful in the future, as large numbers of scientific studies continue to be published detailing clear associations between specific genetic variants and disease.[66][67]
References
- ↑ Alberry, M., et al. (2007). "Free fetal DNA in maternal plasma in anembryonic pregnancies: confirmation that the origin is the trophoblast". Prenatal Diagnosis 27 (5): 415-418.
- ↑ Gupta, A. K., et al. (2004). "Detection of fetal DNA and RNA in placenta-derived syncytiotrophoblast microparticles generated in vitro". Clinical Chemistry 50 (11): 2187-2190.
- ↑ Wang, Eric; Batey, Annette; Struble, Craig; Musci, Thomas; Song, Ken; Oliphant, Arnold (July 2013). "Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma". Prenatal Diagnosis. 33 (7): 662–666. doi:10.1002/pd.4119. PMID 23553731.
- ↑ Smets, E. M.; Visser, A.; Go, A. T.; van Vugt, J. M.; Oudejans, C. B. (2006). "Novel biomarkers in preeclampsia". Clinica Chimica Acta: International Journal of Clinical Chemistry 364 (1-2): 22-32.
- ↑ Lo, Y. M., et al. (1998). "Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis". American Journal of Human Genetics. 62 (4): 768-775.
- ↑ Lo, Y. M., et al. (1999). "Rapid clearance of fetal DNA from maternal plasma". American Journal of Human Genetics 64 (1): 218-224.
- ↑ Chan, K. C., et al. (2004). "Size distributions of maternal and fetal DNA in maternal plasma". Clinical Chemistry 50 (1): 88-92.
- 1 2 Li, Y., et al. (2004). "Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms". Clinical Chemistry 50 (6): 1002-1011.
- ↑ Li, Y., et al. (2005). "Detection of paternally inherited fetal point mutations for beta-thalassemia using size-fractionated cell-free DNA in maternal plasma". The Journal of the American Medical Association 293 (7): 843-849.
- 1 2 Hahn, S.; Chitty, L. S. (2008). "Noninvasive prenatal diagnosis: current practice and future perspectives". Current Opinion in Obstetrics & Gynecology 20 (2): 146-151.
- ↑ Fan, H. C.; Blumenfeld, Y. J.; Chitkara, U.; Hudgins, L.; Quake, S. R. (2008). "Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood". Proceedings of the National Academy of Sciences of the United States of America 105 (42): 16266-16271.
- 1 2 Lo, Y. M., et al. (2007). "Digital PCR for the molecular detection of fetal chromosomal aneuploidy". Proceedings of the National Academy of Sciences of the United States of America 104 (32): 13116-13121.
- ↑ Lo, Y. M., et al. (1998). "Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma". The New England Journal of Medicine 339 (24): 1734-1738.
- ↑ Allyse, M.; Sayres, L. C.; King, J. S.; Norton, M. E.; Cho, M. K. (2012). "Cell-free fetal DNA testing for fetal aneuploidy and beyond: clinical integration challenges in the US context". Human Reproduction 27 (11): 3123-3131.
- ↑ Mujezinovic, F.; Alfirevic, Z. (2007). "Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review". Obstetrics and Gynecology 110 (3): 687-694.
- 1 2 Lo, Y. M. (2008). "Fetal nucleic acids in maternal plasma". Annals of the New York Academy of Sciences 1137: 140-143.
- ↑ The NHS RAPID project | Reliable Accurate Prenatal non-Invasive Diagnosis
- ↑ Guibert, J., et al. (2003). "Kinetics of SRY gene appearance in maternal serum: detection by real time PCR in early pregnancy after assisted reproductive technique". Human Reproduction 18 (8): 1733-1736.
- 1 2 3 4 5 6 Wright, C. F.; Burton, H. (2009). "The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis". Human Reproduction Update 15 (1): 139-151.
- ↑ Chiu, R. W., et al. (2001). "Effects of blood-processing protocols on fetal and total DNA quantification in maternal plasma". Clinical Chemistry 47 (9): 1607-1613.
- ↑ Legler, T. J., et al. (2007). "Workshop report on the extraction of foetal DNA from maternal plasma". Prenatal Diagnosis 27 (9): 824-829.
- ↑ Dhallan, R., et al. (2004). "Methods to increase the percentage of free fetal DNA recovered from the maternal circulation". The Journal of the American Medical Association 291 (9): 1114-1119.
- ↑ Benachi, A., et al. (2005). "Impact of formaldehyde on the in vitro proportion of fetal DNA in maternal plasma and serum". Clinical Chemistry 51 (1): 242-244.
- ↑ Chinnapapagari, S. K.; Holzgreve, W.; Lapaire, O.; Zimmermann, B.; Hahn, S. (2005). "Treatment of maternal blood samples with formaldehyde does not alter the proportion of circulatory fetal nucleic acids (DNA and mRNA) in maternal plasma". Clinical Chemistry 51 (3): 652-655.
- 1 2 Traeger-Synodinos, J. (2006). "Real-time PCR for prenatal and preimplantation genetic diagnosis of monogenic diseases". Molecular Aspects of Medicine 27 (2-3): 176-191.
- 1 2 Al-Yatama, M. K., et al. (2001). "Detection of Y chromosome-specific DNA in the plasma and urine of pregnant women using nested polymerase chain reaction". Prenatal Diagnosis 21 (5): 399-402.
- ↑ Boon, E. M., et al. (2007). "Y chromosome detection by Real Time PCR and pyrophosphorolysis-activated polymerisation using free fetal DNA isolated from maternal plasma". Prenatal Diagnosis 27 (10): 932-937.
- ↑ Hill, M., et al. (2010). "Steroid metabolome in fetal and maternal body fluids in human late pregnancy". The Journal of Steroid Biochemistry and Molecular Biology 122 (4): 114-132.
- ↑ Pierce, K. E.; Rice, J. E.; Sanchez, J. A.; Brenner, C.; Wangh, L. J. (2000). "Real-time PCR using molecular beacons for accurate detection of the Y chromosome in single human blastomeres". Molecular Human Reproduction 6 (12): 1155-1164.
- ↑ Rice, J. E.; Sanchez, J. A.; Pierce, K. E.; Wangh, L. J. (2002). "Real-time PCR with molecular beacons provides a highly accurate assay for detection of Tay-Sachs alleles in single cells". Prenatal Diagnosis 22 (12): 1130-1134.
- ↑ Pierce, K. E.; Rice, J. E.; Sanchez, J. A.; Wangh, L. J. (2003). "Detection of cystic fibrosis alleles from single cells using molecular beacons and a novel method of asymmetric real-time PCR". Molecular Human Reproduction 9 (12): 815-820.
- ↑ Vrettou, C., et al. (2004). "Real-time PCR for single-cell genotyping in sickle cell and thalassemia syndromes as a rapid, accurate, reliable, and widely applicable protocol for preimplantation genetic diagnosis". Human Mutation 23 (5): 513-521.
- 1 2 Zimmermann, B. G., et al. (2008). "Digital PCR: a powerful new tool for noninvasive prenatal diagnosis?". Prenatal Diagnosis 28 (12): 1087-1093.
- ↑ Quake, S. (2007). "At the interface of physics and biology". BioTechniques 43 (1): 19.
- ↑ Chiu, R. W.; Lo, Y. M. (2010). "Pregnancy-associated microRNAs in maternal plasma: a channel for fetal-maternal communication?". Clinical Chemistry 56 (11): 1656-1657.
- 1 2 Ding, C. (2008). "Maldi-TOF mass spectrometry for analyzing cell-free fetal DNA in maternal plasma". Methods in Molecular Biology 444: 253-267.
- ↑ Akolekar, R.; Farkas, D. H.; VanAgtmael, A. L.; Bombard, A. T.; Nicolaides, K. H. (2010). "Fetal sex determination using circulating cell-free fetal DNA (ccffDNA) at 11 to 13 weeks of gestation". Prenatal Diagnosis 30 (10): 918-923.
- ↑ Tong, Y. K.; Chiu, R. W.; Chan, K. C.; Leung, T. Y.; Lo, Y. M. (2012). "Technical concerns about immunoprecipitation of methylated fetal DNA for noninvasive trisomy 21 diagnosis". Nature Medicine 18 (9): 1327-1328; author reply 1328-1329.
- ↑ Papageorgiou, E. A., et al. (2011). "Fetal-specific DNA methylation ratio permits noninvasive prenatal diagnosis of trisomy 21". Nature Medicine 17 (4): 510-513.
- 1 2 White, H. E., Dent, C. L.; Hall, V. J.; Crolla, J. A.; Chitty, L. S. (2012). "Evaluation of a novel assay for detection of the fetal marker RASSF1A: facilitating improved diagnostic reliability of noninvasive prenatal diagnosis". PloS One 7 (9): e45073.
- ↑ Ng, E. K., et al. (2002). "Presence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individuals". Clinical Chemistry 48 (8): 1212-1217.
- ↑ Baird, P. A.; Anderson, T. W.; Newcombe, H. B.; Lowry, R. B. (1988). "Genetic disorders in children and young adults: a population study". American Journal of Human Genetics 42 (5): 677-693.
- ↑ Scheffer, P. G., et al. (2010). "Reliability of fetal sex determination using maternal plasma". Obstetrics and Gynecology 115 (1): 117-126.
- 1 2 3 Bustamante-Aragones, A.; Gonzalez-Gonzalez, C.; de Alba, M. R.; Ainse, E.; Ramos, C. (2010). "Noninvasive prenatal diagnosis using ccffDNA in maternal blood: state of the art". Expert Review of Molecular Diagnostics 10 (2): 197-205.
- ↑ Zimmermann, B.; El-Sheikhah, A.; Nicolaides, K.; Holzgreve, W.; Hahn, S. (2005). "Optimized real-time quantitative PCR measurement of male fetal DNA in maternal plasma". Clinical Chemistry 51 (9): 1598-1604.
- 1 2 Finning, K. M.; Chitty, L. S. (2008). "Non-invasive fetal sex determination: impact on clinical practice". Seminars in Fetal & Neonatal Medicine 13 (2): 69-75.
- ↑ Markey, C. M.; Wadia, P. R.; Rubin, B. S.; Sonnenschein, C; Soto, A. M. (2005). "Long-term effects of fetal exposure to low doses of the xenoestrogen bisphenol-A in the female mouse genital tract". Biology of Reproduction 72 (6): 1344-1351.
- ↑ Sayres, L. C.; Cho, M. K. (2011). "Cell-free fetal nucleic acid testing: a review of the technology and its applications". Obstetrical & Gynecological Survey 66 (7): 431-442.
- ↑ Hill, M.; Barrett, A. N.; White, H.; Chitty, L. S. (2012). "Uses of cell free fetal DNA in maternal circulation". Best Practice & Research: Clinical Obstetrics & Gynaecology 26 (5): 639-654.
- ↑ Norbury, G.; Norbury, C. J. (2008). "Non-invasive prenatal diagnosis of single gene disorders: how close are we?". Seminars in Fetal & Neonatal Medicine 13 (2): 76-83.
- 1 2 Li, Y.; Page-Christiaens, G. C.; Gille, J. J.; Holzgreve, W.; Hahn, S. (2007). "Non-invasive prenatal detection of achondroplasia in size-fractionated cell-free DNA by MALDI-TOF MS assay". Prenatal Diagnosis 27 (1): 11-17.
- ↑ De Die-Smulders, C. E. M.; De Wert, G. M. W. R.; Liebaers, I.; Tibben, A.; Evers-Kiebooms, G. (2013). "Reproductive options for prospective parents in families with Huntington's disease: Clinical, psychological and ethical reflections". Human Reproduction Update. 19 (3): 304–315. doi:10.1093/humupd/dms058. PMID 23377865.
- ↑ Chinen, P. A., et al. (2010). "Noninvasive determination of fetal rh blood group, D antigen status by cell-free DNA analysis in maternal plasma: experience in a Brazilian population". American Journal of Perinatology 27 (10): 759-762.
- ↑ Kolialexi, A.; Tounta, G.; Mavrou, A. (2010). "Noninvasive fetal RhD genotyping from maternal blood". Expert Review of Molecular Diagnostics 10 (3): 285-296.
- 1 2 Moise, K. J. (2008). "Fetal anemia due to non-Rhesus-D red-cell alloimmunization". Seminars in Fetal & Neonatal Medicine 13 (4): 207-214.
- ↑ Lo, Y. M., et al. (1999). "Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma". Cancer Research 59 (21): 5452-5455.
- ↑ Wataganara, T., et al. (2003). "Maternal serum cell-free fetal DNA levels are increased in cases of trisomy 13 but not trisomy 18". Human Genetics 112 (2): 204-208.
- ↑ Chiu, R. W.; Lo, Y. M. (2011). "Non-invasive prenatal diagnosis by fetal nucleic acid analysis in maternal plasma: the coming of age". Seminars in Fetal & Neonatal Medicine 16 (2): 88-93.
- ↑ Mersy, E.; Smits, L. J. M.; Van Winden, L. A. A. P.; De Die-Smulders, C. E. M.; Paulussen, A. D. C.; MacVille, M. V. E.; Coumans, A. B. C.; Frints, S. G. M. (2013). "Noninvasive detection of fetal trisomy 21: Systematic review and report of quality and outcomes of diagnostic accuracy studies performed between 1997 and 2012". Human Reproduction Update. 19 (4): 318–29. doi:10.1093/humupd/dmt001. PMID 23396607.
- ↑ Noninvasive Prenatal Diagnosis of Fetal Aneuploidy Using Cell-Free Fetal Nucleic Acids in Maternal Blood: Clinical Policy (Effective 05/01/2013) from Oxford Health Plans
- ↑ Bianchi, D. W.; Parker, R. L.; Wentworth, J.; Madankumar, R.; Saffer, C.; Das, A. F.; Craig, J. A.; Chudova, D. I.; Devers, P. L.; Jones, K. W.; Oliver, K.; Rava, R. P.; Sehnert, A. J. (2014). "DNA Sequencing versus Standard Prenatal Aneuploidy Screening". New England Journal of Medicine. 370 (9): 799–808. doi:10.1056/NEJMoa1311037. PMID 24571752.. A recent study in the New England Journal of Medicine demonstrated the feasibility of using NIPT in a low risk population.
- ↑ Lo, Y. M., et al. (1999). "Increased fetal DNA concentrations in the plasma of pregnant women carrying fetuses with trisomy 21". Clinical Chemistry 45 (10): 1747-1751.
- ↑ Leung, T. N.; Zhang, J.; Lau, T. K.; Chan, L. Y.; Lo, Y. M. (2001). "Increased maternal plasma fetal DNA concentrations in women who eventually develop preeclampsia". Clinical Chemistry 47 (1): 137-139.
- ↑ Zhong, X. Y.; Holzgreve, W.; Hahn, S. (2002). "The levels of circulatory cell free fetal DNA in maternal plasma are elevated prior to the onset of preeclampsia". Hypertension in Pregnancy: Official Journal of the International Society for the Study of Hypertension in Pregnancy 21 (1): 77-83.
- ↑ Yurkiewicz, I. R.; Korf, B. R.; Lehmann, L. S. (2014). "Prenatal whole-genome sequencing--is the quest to know a fetus's future ethical?". New England Journal of Medicine. 370 (3): 195–7. doi:10.1056/NEJMp1215536. PMID 24428465.
- ↑ Wellcome Trust Case Control Consortium; Clayton, David G.; Cardon, Lon R.; Craddock, Nick; Deloukas, Panos; Duncanson, Audrey; Kwiatkowski, Dominic P.; McCarthy, Mark I.; Ouwehand, Willem H.; Samani, Nilesh J.; Todd; Donnelly, Peter; Barrett, Jeffrey C.; Burton, Paul R.; Davison, Dan; Donnelly, Peter; Easton, Doug; Evans, David; Leung, Hin-Tak; Marchini, Jonathan L.; Morris; Spencer, Chris C. A.; Tobin, Martin D.; Cardon, Lon R.; Clayton, David G.; Attwood, Antony P.; Boorman, James P.; Cant, Barbara; Everson, Ursula; Hussey, Judith M. (June 2007). "Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls". Nature. 447 (7145): 661–78. Bibcode:2007Natur.447..661B. doi:10.1038/nature05911. PMC 2719288
 . PMID 17554300.
. PMID 17554300. - ↑ Mailman MD; Feolo M; Jin Y; Kimura M; Tryka K; Bagoutdinov R; Hao L; Kiang A; Paschall J; Phan L; Popova N; Pretel S; Ziyabari L; Lee M; Shao Y; Wang ZY; Sirotkin K; Ward M; Kholodov M; Zbicz K; Beck J; Kimelman M; Shevelev S; Preuss D; Yaschenko E; Graeff A; Ostell J; Sherry ST (October 2007). "The NCBI dbGaP database of genotypes and phenotypes". Nat. Genet. 39 (10): 1181–6. doi:10.1038/ng1007-1181. PMC 2031016
 . PMID 17898773.
. PMID 17898773.