Caenorhabditis elegans
| Caenorhabditis elegans | |
|---|---|
 | |
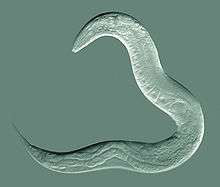
| An adult hermaphrodite C. elegans worm | |
| Scientific classification | |
| Kingdom: | Animalia |
| Phylum: | Nematoda |
| Class: | Chromadorea |
| Order: | Rhabditida |
| Family: | Rhabditidae |
| Genus: | Caenorhabditis |
| Species: | C. elegans |
| Binomial name | |
| Caenorhabditis elegans (Maupas, 1900)[1] | |
Caenorhabditis elegans (/ˌseɪnoʊræbˈdaɪtəs ˈɛləɡænz/[2]) is a free-living (not parasitic), transparent nematode (roundworm), about 1 mm in length,[3] that lives in temperate soil environments. The name is a blend of the Greek caeno- (recent), rhabditis (rod-like)[4] and Latin elegans (elegant). In 1900, Maupas initially named it Rhabditides elegans, Osche placed it in the subgenus Caenorhabditis in 1952, and in 1955, Dougherty raised it to the status of genus.[5]
C. elegans is an unsegmented pseudocoelomate, and lacks a respiratory and a circulatory system. It possesses gut granules which emit a brilliant blue fluorescence, a wave of which is seen at death in a 'death fluorescence'.[6] The majority of these nematodes are hermaphrodites. Males have specialised tails for mating that include spicules.
In 1963, Sydney Brenner proposed research into C. elegans primarily in the area of neuronal development. In 1974, he began research into the molecular and developmental biology of C. elegans, which has since been extensively used as a model organism.[7]
C. elegans was the first multicellular organism to have its whole genome sequenced, and as of 2012, the only organism to have its connectome (neuronal "wiring diagram") completed.[8][9][10]
Anatomy

C. elegans is unsegmented, vermiform, and bilaterally symmetrical. It has a cuticle (a tough outer covering), four main epidermal cords, and a fluid-filled pseudocoelom (body cavity). It also has some of the same organ systems as larger animals. About one in a thousand individuals is male and the rest are hermaphrodites.[11] The basic anatomy of C. elegans includes a mouth, pharynx, intestine, gonad, and collagenous cuticle. Like all nematodes, they have neither a circulatory nor a respiratory system. The four bands of muscles that run the length of the body are connected to a neural system that allows the muscles to move the animal's body only as dorsal bending or ventral bending, but not left or right, except for the head, where the four muscle quadrants are wired independently from one another. When a wave of dorsal/ventral muscle contractions proceeds from the back to the front of the animal, the animal is propelled backwards. When wave of contractions is initiated at the front and proceeds posteriorly along the body, the animal is propelled forwards. Because of this dorsal/ventral bias in body bends, any normal living, moving individual tends to lie on either its left side or its right side when observed crossing a horizontal surface. A set of ridges on the lateral sides of the body cuticle, the alae, are believed to give the animal added traction during these bending motions.
The pharynx is a muscular food pump in the head of C. elegans, which is triangular in cross-section. This grinds food and transports it directly to the intestine. A set of "valve cells" connects the pharynx to the intestine, but how this valve operates is not understood. After digestion, the contents of the intestine are released via the rectum, as is the case with all other nematodes. No direct connection exists between the pharynx and the excretory canal, which functions in the release of liquid urine.
Males have a single-lobed gonad, a vas deferens, and a tail specialized for mating, which incorporates spicules. Hermaphrodites have two ovaries, oviducts, spermatheca, and a single uterus.
Microanatomy
Numerous gut granules are present in the intestine of C. elegans, the functions of which are still not fully known, as are many other aspects of this nematode, despite the many years that it has been studied. These gut granules are found in all of the Rhabditida orders. They are very similar to lysosomes in that they feature an acidic interior and the capacity for endocytosis, but they are considerably larger, reinforcing the view of their being storage organelles. A remarkable feature of the granules is that when they are observed under ultraviolet light, they react by emitting an intense blue fluorescence. Another phenomenon seen is termed 'death fluorescence'. As the worms die, a dramatic burst of blue fluorescence is emitted. This death fluorescence typically takes place in an anterior to posterior wave that moves along the intestine, and is seen in both young and old worms, whether subjected to lethal injury or peacefully dying of old age. Many theories have been posited on the functions of the gut granules, with earlier ones being eliminated by later findings. They are thought to store zinc as one of their functions. Recent chemical analysis has identified the blue fluorescent material they contain as a glycosylated form of anthranilic acid (AA). The need for the large amounts of AA the many gut granules contain is questioned. One possibility is that the AA is antibacterial and used in defense against invading pathogens. Another possibility is that the granules provide photoprotection: the bursts of AA fluorescence entail the conversion of damaging UV light to relatively harmless visible light. This is seen a possible link to the melanin–containing melanosomes.[12]
Reproduction and development


All cells of the germline arise from a single primordial germ cell, called the P4 cell established early in embryogenesis.[14][15] This germ cell divides to generate two further germ cells and these do not divide further until after hatching.[15] The hermaphrodite, which is considered to be a specialized form of self-fertile female because its soma is female whereas its germline produces male gametes first, lays eggs through its uterus after internal fertilization. Under environmental conditions which are favourable for reproduction, hatched larvae develop through four stages or molts, designated as L1 to L4. When conditions are stressed as in food insufficiency, C. elegans can enter an alternative third larval stage called the dauer state. Dauer is German for permanent. Dauer larvae are stress-resistant; they are thin and their mouths are sealed and cannot take in food, and they can remain in this stage for a few months.[16] Hermaphrodites produce all their sperm in the L4 stage (150 sperm per gonadal arm) and then produce only oocytes. The sperm cells are stored in the same area of the gonad as the oocytes until the first oocyte pushes the sperm into the spermatheca (a chamber wherein the oocytes become fertilized by the sperm).[17]
The male can inseminate the hermaphrodite, which will preferentially use male sperm (both types of sperm are stored in the spermatheca). The sperm of C. elegans is ameboid, lacking flagella and acrosomes.[18] When self-inseminated, the wild-type worm will lay about 300 eggs. When inseminated by a male, the number of progeny can exceed 1,000. At 20 °C, the laboratory strain of C. elegans (N2) has an average lifespan around 2–3 weeks and a generation time around 4 days.
Nematodes have a fixed, genetically determined number of cells, a phenomenon known as eutely. The male C. elegans has 1031 cells, a number which does not change after cell division ceases at the end of the larval period. Growth is solely due to an increase in the size of individual cells.[19]
C. elegans has five pairs of autosomes and one pair of sex chromosomes. Sex in C. elegans is based on an X0 sex-determination system. Hermaphrodites of C. elegans have a matched pair of sex chromosomes (XX); the rare males have only one sex chromosome (X0).

Ecology
The different Caenorhabditis species occupy various nutrient- and bacteria-rich environments. They feed on the bacteria that develop in decaying organic matter. Soil lacks enough organic matter to support self-sustaining populations. C. elegans can survive on a diet of a variety of many kinds of bacteria, but its wild ecology is largely unknown. Most laboratory strains were taken from artificial environments such as gardens and compost piles. More recently, C. elegans has been found to thrive in other kinds of organic matter, particularly rotting fruit.[20] Invertebrates such as millipedes, insects, isopods, and gastropods can transport dauer larvae to various suitable locations. The larvae have also been seen to feed on their hosts when they die.[21] Nematodes can survive desiccation, and in C. elegans, the mechanism for this capability has been demonstrated to be late embryogenesis abundant proteins.[22] C. elegans, as other nematodes, can be eaten by predator nematodes and other omnivores, including some insects.[23]
Research use
In 1963, Sydney Brenner proposed using C. elegans as a model organism for the investigation primarily of neural development in animals. It is one of the simplest organisms with a nervous system. In the hermaphrodite, this system comprises 302 neurons[24] the pattern of which has been comprehensively mapped, in what is known as a connectome, and shown to be a small-world network.[25] Research has explored the neural and molecular mechanisms that control several behaviors of C. elegans, including chemotaxis, thermotaxis, mechanotransduction, learning, memory, and mating behaviour.[26] Brenner also chose it as it is easy to grow in bulk populations, and convenient for genetic analysis.[27] It is a multicellular eukaryotic organism, yet is simple enough to be studied in great detail. The transparency of C. elegans facilitates the study of cellular differentiation and other developmental processes in the intact organism. The spicules in the male clearly distinguish males from females. Strains are cheap to breed and can be frozen. When subsequently thawed, they remain viable, allowing long-term storage.[28]
Notable findings
The developmental fate of every single somatic cell (959 in the adult hermaphrodite; 1031 in the adult male) has been mapped.[29][30] These patterns of cell lineage are largely invariant between individuals, whereas in mammals, cell development is more dependent on cellular cues from the embryo. The first cell divisions of early embryogenesis in C. elegans are among the best understood examples of asymmetric cell divisions.[31]
Programmed cell death (apoptosis) eliminates many additional cells (131 in the hermaphrodite, most of which would otherwise become neurons); this "apoptotic predictability" has contributed to the elucidation of some apoptotic genes. Cell death-promoting genes and a single cell-death inhibitor have been identified.[32]

RNA interference (RNAi) is a relatively straightforward method of disrupting the function of specific genes. Silencing the function of a gene can sometimes allow a researcher to infer its possible function(s). The nematode can be soaked in, injected with, or fed with genetically transformed bacteria that express the double-stranded RNA of interest, the sequence of which complements the sequence of the gene that the researcher wishes to disable.[33] RNAi has emerged as a powerful tool in the study of functional genomics. In C. elegans, it has been used to analyse gene functions and the report claims the promise of future findings in the systematic genetic interactions.[34]
Environmental RNAi uptake is much worse in other species of worms in the Caenorhabditis genus. Although injecting RNA into the body cavity of the animal induces gene silencing in most species, only C. elegans and a few other distantly related nematodes can take up RNA from the bacteria they eat for RNAi.[35] This ability has been mapped down to a single gene, sid-2, which, when inserted as a transgene in other species, allows them to take up RNA for RNAi as C. elegans does.[36]
Research into meiosis has been considerably simplified since every germ cell nucleus is at the same given position as it moves down the gonad, so is at the same stage in meiosis. In an early phase of meiosis, the oocytes become extremely resistant to radiation and this resistance depends on expression of genes rad51 and atm that have key roles in recombinational repair.[37][38] Gene mre-11 also plays a crucial role in recombinational repair of DNA damage during meiosis.[39] A study of the frequency of outcrossing in natural populations showed that selfing is the predominant mode of reproduction in C. elegans, but that infrequent outcrossing events occur at a rate around 1%.[40] Meioses that result in selfing are unlikely to contribute significantly to beneficial genetic variability, but these meioses may provide the adaptive benefit of recombinational repair of DNA damages that arise, especially under stressful conditions.[41]
Nicotine dependence can also be studied using C. elegans because it exhibits behavioral responses to nicotine that parallel those of mammals. These responses include acute response, tolerance, withdrawal, and sensitization.[42]
As for most model organisms, scientists that work in the field curate a dedicated online database and the WormBase is that for C. elegans. The WormBase attempts to collate all published information on C. elegans and other related nematodes. Their website has advertised a reward of $4000 for the finder of a new species of closely related nematode.[43] Such a discovery would broaden research opportunities with the worm.[44]
C. elegans has been a model organism for research into ageing; for example, the inhibition of an insulin-like growth factor signaling pathway has been shown to increase adult lifespan threefold.[45] Moreover, extensive research on C. elegans has identified RNA-binding proteins as essential factors during germline and early embryonic development.[46]
C. elegans is notable in animal sleep studies as the most primitive organism to display sleep-like states. In C. elegans, a lethargus phase occurs shortly before each moult.[47]
Spaceflight research
C. elegans made news when specimens were discovered to have survived the Space Shuttle Columbia disaster in February 2003.[48] Later, in January 2009, live samples of C. elegans from the University of Nottingham were announced to be spending two weeks on the International Space Station that October, in a space research project to explore the effects of zero gravity on muscle development and physiology. The research was primarily about genetic basis of muscle atrophy, which relates to spaceflight or being bed-ridden, geriatric, or diabetic.[49] Descendants of the worms aboard Columbia in 2003 were launched into space on Endeavour for the STS-134 mission.[50]
Genome
C. elegans was the first multicellular organism to have its whole genome sequenced. The sequence was published in 1998,[51] although some small gaps were present; the last gap was finished by October 2002.
Size and gene content. The C. elegans genome is about 100 million base pairs long and consists of six chromosomes and a mitochondrial genome. Its gene density is about one gene per five kilo-base pairs. Introns make up 26% and intergenic regions 47% of the genome. Many genes are arranged in clusters and how many of these are operons is unclear.[52] C. elegans and other nematodes are among the few eukaryotes currently known to have operons; these include trypanosomes, flatworms (notably the trematode Schistosoma mansoni), and a primitive chordate tunicate Oikopleura dioica. Many more organisms are likely to be shown to have these operons.[53]
Protein-coding genes. The genome contains an estimated 20,470 protein-coding genes.[54] About 35% of C. elegans genes have human homologs. Remarkably, human genes have been shown repeatedly to replace their C. elegans homologs when introduced into C. elegans. Conversely, many C. elegans genes can function similarly to mammalian genes.[16] The number of known RNA genes in the genome has increased greatly due to the 2006 discovery of a new class of 21U-RNA genes,[55] and the genome is now believed to contain more than 16,000 RNA genes, up from as few as 1,300 in 2005.[56] Scientific curators continue to appraise the set of known genes; new gene models continue to be added and incorrect ones modified or removed.
Related genomes. In 2003, the genome sequence of the related nematode C. briggsae was also determined, allowing researchers to study the comparative genomics of these two organisms.[57] The genome sequences of more nematodes from the same genus e.g., C. remanei,[58] C. japonica[59] and C. brenneri (named after Brenner), have also been studied using the shotgun sequencing technique.[60] These sequences have now been completed.[61][62]
The reference C. elegans genome sequence continues to change as new evidence reveals errors in the original sequencing. Most changes are minor, adding or removing only a few base pairs of DNA. For example, the WS202 release of WormBase (April 2009) added two base pairs to the genome sequence.[63] Sometimes, more extensive changes are made as noted in the WS197 release of December 2008, which added a region of over 4,300 bp to the sequence.[64][65]
Scientific community
In 2002, the Nobel Prize in Physiology or Medicine was awarded to Sydney Brenner, H. Robert Horvitz, and John Sulston for their work on the genetics of organ development and programmed cell death in C. elegans. The 2006 Nobel Prize in Physiology or Medicine was awarded to Andrew Fire and Craig C. Mello for their discovery of RNA interference in C. elegans.[66] In 2008, Martin Chalfie shared a Nobel Prize in Chemistry for his work on green fluorescent protein; some of the research involved the use of C. elegans.
Many scientists who research C. elegans closely connect to Sydney Brenner, with whom almost all research in this field began in the 1970s; they have worked as either a postdoctoral or a postgraduate researcher in Brenner's lab or in the lab of someone who previously worked with Brenner. Most who worked in his lab later established their own worm research labs, thereby creating a fairly well-documented "lineage" of C. elegans scientists, which was recorded into the WormBase database in some detail at the 2003 International Worm Meeting.[67]
See also
| Wikimedia Commons has media related to Caenorhabditis elegans. |
References
- ↑ Maupas, É (1900). "Modes et formes de reproduction des nématodes". Archives de Zoologie Expérimentale et Générale. 8: 463–624.
- ↑ "Caenorhabditis". Merriam-Webster Dictionary.
- ↑ Wood, WB (1988). The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press. p. 1. ISBN 0-87969-433-5.
- ↑ καινός (caenos) = new, recent; ῥάβδος (rhabdos) = rod, wand.
- ↑ Ferris, H (30 November 2013). "Caenorhabditis elegans". University of California, Davis. Retrieved 2013-11-19.
- ↑ Coburn, C.; Allman, E.; Mahanti, P.; Benedetto, A.; Cabreiro, F.; Pincus, Z.; Matthijssens, F.; Araiz, C.; Mandel, A.; Vlachos, M.; Edwards, S. A.; Fischer, G.; Davidson, A.; Pryor, R. E.; Stevens, A.; Slack, F. J.; Tavernarakis, N.; Braeckman, B. P.; Schroeder, F. C.; Nehrke, K.; Gems, D. (2013). "Anthranilate Fluorescence Marks a Calcium-Propagated Necrotic Wave That Promotes Organismal Death in C. Elegans". PLoS Biology. 11 (7): e1001613. doi:10.1371/journal.pbio.1001613. PMC 3720247
 . PMID 23935448.
. PMID 23935448. 
- ↑ Brenner, S (1974). "The Genetics of Caenorhabditis elegans". Genetics. 77 (1): 71–94. PMC 1213120
 . PMID 4366476.
. PMID 4366476. - ↑ White, John Graham; et al. (1986). "The structure of the nervous system of the nematode Caenorhabditis elegans". Philos. Trans. R. Soc. Lond., B, Biol. Sci. 314 (1165): 1–340. Bibcode:1986RSPTB.314....1W. doi:10.1098/rstb.1986.0056. PMID 22462104.
- ↑ White, John Graham (2013). "Getting into the mind of a worm—a personal view". WormBook: 1–10. doi:10.1895/wormbook.1.158.1. PMC 4781474
 . PMID 23801597.
. PMID 23801597. - ↑ Jabr, Ferris (2012-10-02). "The Connectome Debate: Is Mapping the Mind of a Worm Worth It?". Scientific American. Retrieved 2014-01-18.
- ↑ Alberts, B; Johnson, A; Lewis, J; Raff, M; Roberts, K; Walter, P (2007). Molecular Biology of the Cell (5th ed.). Garland Science. p. 1321. ISBN 978-0-8153-4105-5.
- ↑ Coburn, C; Gems, D (2013). "The mysterious case of the C. Elegans gut granule: Death fluorescence, anthranilic acid and the kynurenine pathway". Frontiers in Genetics. 4: 151. doi:10.3389/fgene.2013.00151. PMC 3735983
 . PMID 23967012.
. PMID 23967012. - ↑ Loer, C. M.; Kenyon, C. J. (1993-12-01). "Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans". The Journal of Neuroscience. 13 (12): 5407–5417. ISSN 0270-6474.
- ↑ Kimble J, Crittenden SL. Germline proliferation and its control. 2005 Aug 15. In: WormBook: The Online Review of C. elegans Biology [Internet]. Pasadena (CA): WormBook; 2005-. Available from: http://www.ncbi.nlm.nih.gov/books/NBK19769/
- 1 2 "WBbt:0006773 (anatomy term)". WormBase (WS242 ed.). May 14, 2014. WBbt:0006773.
- 1 2 "Introduction to C. Elegans". C. Elegans as a model organism. Rutgers University. Archived from the original on 2002-08-18. Retrieved August 15, 2014.
- ↑ Nayak, S; Goree, J; Schedl, T (2004). "fog-2 and the Evolution of Self-Fertile Hermaphroditism in Caenorhabditis". PLoS Biology. 3 (1): e6. doi:10.1371/journal.pbio.0030006. PMC 539060
 . PMID 15630478.
. PMID 15630478. 
- ↑ Ma, X.; Zhao, Y; et al. (Oct 2012). "Transformation: how do nematode sperm become activated and crawl?". Protein Cell. 3 (3 (10)): 755–61. doi:10.1007/s13238-012-2936-2. PMID 22903434.
- ↑ Ruppert, Edward E.; Fox, Richard, S.; Barnes, Robert D. (2004). Invertebrate Zoology (7th ed.). Cengage Learning. p. 753. ISBN 978-81-315-0104-7.
- ↑ Félix, MA; Braendle, C (2010). "The natural history of Caenorhabditis elegans". Current Biology. 20 (22): R965–R969. doi:10.1016/j.cub.2010.09.050. PMID 21093785.
- ↑ Kiontke, K; Sudhaus, W (2006). "Ecology of Caenorhabditis species". WormBook: 1–14. doi:10.1895/wormbook.1.37.1. PMID 18050464.
- ↑ Gal, TZ; Glazer, I; Koltai, H (2004). "An LEA group 3 family member is involved in survival of C. elegans during exposure to stress". FEBS Letters. 577 (1–2): 21–26. doi:10.1016/j.febslet.2004.09.049. PMID 15527756.
- ↑ Elaine R. Ingham Soil biology primer USDA
- ↑ Kosinski, RA; Zaremba, M (2007). "Dynamics of the Model of the Caenorhabditis elegans Neural Network". Acta Physica Polonica B. 38 (6): 2201. Bibcode:2007AcPPB..38.2201K.
- ↑ Watts, DJ; Strogatz, SH (1998). "Collective dynamics of 'small-world' networks". Nature. 393 (6684): 440–442. Bibcode:1998Natur.393..440W. doi:10.1038/30918. PMID 9623998.
- ↑ Schafer, W.R. (2005). "Deciphering the neural and molecular mechanisms of C. elegans behaviour". Curr.Biol. 15 (17): R723–9. doi:10.1016/j.cub.2005.08.020. PMID 16139205.
- ↑ Avery, L. "Sydney Brenner". Southwestern Medical Center. Archived from the original on August 15, 2011. Alt. URL
- ↑ Brenner, S (1974). "The Genetics of Caenorhabditis elegans". Genetics. 77 (1): 71–94. PMC 1213120
 . PMID 4366476.
. PMID 4366476. - ↑ Sulston, JE; Horvitz, HR (1977). "Post-embryonic cell lineages of the nematode, Caenorhabditis elegans". Developmental Biology. 56 (1): 110–56. doi:10.1016/0012-1606(77)90158-0. PMID 838129.
- ↑ Kimble, J; Hirsh, D (1979). "The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans". Developmental Biology. 70 (2): 396–417. doi:10.1016/0012-1606(79)90035-6. PMID 478167.
- ↑ Gönczy, P (2005). "Asymmetric cell division and axis formation in the embryo". WormBook: 1–20. doi:10.1895/wormbook.1.30.1.
- ↑ Peden, E (Aug 2008). "Cell death specification in C. elegans". Cell Cycle. 7 (16): 2479–2484. doi:10.4161/cc.7.16.6479. PMC 2651394
 . PMID 18719375.
. PMID 18719375. - ↑ Kamath, RS; et al. (2003). "Systematic functional analysis of the Caenorhabditis elegans genome using RNAi". Nature. 421 (6920): 231–237. Bibcode:2003Natur.421..231K. doi:10.1038/nature01278. PMID 12529635.
- ↑ Fortunato, AI & Fraser, AG (2005). "Uncover genetic interaction in Caenorhabditis elegans by RNA interference". Biosci Rep. 25 (Oct-Dec (5-6)): 299–307. doi:10.1007/s10540-005-2892-7. PMID 16307378.
- ↑ Félix, M-A (2008). "RNA interference in nematodes and the chance that favored Sydney Brenner". Journal of Biology. 7 (9): 34–56. doi:10.1186/jbiol97. PMC 2776389
 . PMID 19014674.
. PMID 19014674. - ↑ Winston, WM; Sutherlin, M; Wright, AJ; Feinberg, EH; Hunter, CP (2007). "Caenorhabditis elegans SID-2 is required for environmental RNA interference". Proceedings of the National Academy of Sciences. 104 (25): 10565–70. Bibcode:2007PNAS..10410565W. doi:10.1073/pnas.0611282104. PMC 1965553
 . PMID 17563372.
. PMID 17563372. - ↑ Takanami T, Mori A, Takahashi H, Higashitani A (November 2000). "Hyper-resistance of meiotic cells to radiation due to a strong expression of a single recA-like gene in Caenorhabditis elegans". Nucleic Acids Res. 28 (21): 4232–6. doi:10.1093/nar/28.21.4232. PMC 113154
 . PMID 11058122.
. PMID 11058122. - ↑ Takanami T, Zhang Y, Aoki H, Abe T, Yoshida S, Takahashi H, Horiuchi S, Higashitani A (September 2003). "Efficient repair of DNA damage induced by heavy ion particles in meiotic prophase I nuclei of Caenorhabditis elegans". J. Radiat. Res. 44 (3): 271–6. doi:10.1269/jrr.44.271. PMID 14646232.
- ↑ Chin GM, Villeneuve AM (March 2001). "C. elegans mre-11 is required for meiotic recombination and DNA repair, but is dispensable for the meiotic G(2) DNA damage checkpoint". Genes Dev. 15 (5): 522–34. doi:10.1101/gad.864101. PMC 312651
 . PMID 11238374.
. PMID 11238374. - ↑ Barrière A, Félix MA (July 2005). "High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations". Curr. Biol. 15 (13): 1176–84. doi:10.1016/j.cub.2005.06.022. PMID 16005289.
- ↑ Bernstein H and Bernstein C (2013). Evolutionary Origin and Adaptive Function of Meiosis. In Meiosis: Bernstein C and Bernstein H, editors. ISBN 978-953-51-1197-9, InTech, http://www.intechopen.com/books/meiosis/evolutionary-origin-and-adaptive-function-of-meiosis
- ↑ Feng, Z; Li, W; Ward, A; Piggott, BJ; Larkspur, ER; Sternberg, PW; Xu, XZ (2006). "A C. elegans model of nicotine-dependent behavior: regulation by TRP family channels". Cell. 127 (3): 621–633. doi:10.1016/j.cell.2006.09.035. PMC 2859215
 . PMID 17081982.
. PMID 17081982. - ↑ "Caenorhabditis isolation guide". WormBase. Archived from the original on November 7, 2007. Retrieved 2007-08-30. Alt. URL
- ↑ Dolgin, E (2007). "Slime for a dime". Science. 317 (5842): 1157. doi:10.1126/science.317.5842.1157b.
- ↑ Wolkow, CA; Kimura, KD; Lee, M-S; Ruvkun, G (2000). "Regulation of C. elegans Life-span by Insulin-like Signaling in the Nervous System". Science. 290 (5489): 147–150. Bibcode:2000Sci...290..147W. doi:10.1126/science.290.5489.147. PMID 11021802.
- ↑ Hanazawa M, Yonetani M & Sugimoto A (Mar 21, 2011). "PGL proteins self associate and bind RNP's to mediate germ granule assembly in C. elegans". Cell Biology. 192 (192(6)): 929–937. doi:10.1083/jcb.201010106. PMC 3063142
 . PMID 21402787.
. PMID 21402787. - ↑ Iwanir, S.; Tramm, N.; et al. (Mar 2013). "The microarchitecture of C. elegans behavior during lethargus: homeostatic bout dynamics, a typical body posture, and regulation by a central neuron.". Sleep. 82 (36 (3)): 385–95. doi:10.5665/Sleep.2456. PMID 23449971.
- ↑ "Worms survived Columbia disaster". BBC News. 1 May 2003. Retrieved 2008-07-11.
- ↑ "University sends worms into space". BBC News. 17 January 2009. Retrieved 2009-07-09.
- ↑ Klotz, I (16 May 2011). "Legacy Space Worms Flying on Shuttle". Discovery News. Retrieved 2011-05-17.
- ↑ The C. elegans Sequencing Consortium (1998). "Genome sequence of the nematode C. elegans: A platform for investigating biology". Science. 282 (5396): 2012–2018. doi:10.1126/science.282.5396.2012. PMID 9851916.
- ↑ Blumenthal, T; et al. (Jun 2002). "A global analysis of Caenorhabditis elegans operons". Nature. 417 (417(6891)): 851–4. Bibcode:2002Natur.417..851B. doi:10.1038/nature00831. PMID 12075352.
- ↑ Blumenthal, T (2004). "Operons in eukaryotes". Briefings in Functional Genomics and Proteomics. 3 (3): 199–211. doi:10.1093/bfgp/3.3.199. PMID 15642184.
- ↑ "WS227 Release Letter". WormBase. 10 August 2011. Retrieved 2013-11-19.
- ↑ Ruby, JG; Jan, C; Player, C; Axtell, MJ; Lee, W; Nusbaum, C; Ge, H; Bartel, DP (2006). "Large-scale Sequencing Reveals 21U-RNAs and Additional MicroRNAs and Endogenous siRNAs in C. elegans". Cell. 127 (6): 1193–207. doi:10.1016/j.cell.2006.10.040. PMID 17174894.
- ↑ Stricklin, SL; Griffiths-Jones, S; Eddy, SR (2005). "C. elegans noncoding RNA genes". WormBook. doi:10.1895/wormbook.1.1.1.
- ↑ Stein, LD; et al. (2003). "The Genome Sequence of Caenorhabditis briggsae: A Platform for Comparative Genomics". PLoS Biology. 1 (2): 166–192. doi:10.1371/journal.pbio.0000045. PMC 261899
 . PMID 14624247.
. PMID 14624247. 
- ↑ Genome Sequencing Center. "Caenorhabditis remanei: Background". Washington University School of Medicine. Archived from the original on 2008-06-16. Retrieved 2008-07-11.
- ↑ Genome Sequencing Center. "Caenorhabditis japonica: Background". Washington University School of Medicine. Archived from the original on 2008-06-26. Retrieved 2008-07-11.
- ↑ Staden, R (1979). "A strategy of DNA sequencing employing computer programs". Nucleic Acids Research. 6 (7): 2601–10. doi:10.1093/nar/6.7.2601. PMC 327874
 . PMID 461197.
. PMID 461197. - ↑ "UCSC genome browser". Retrieved 8 July 2014.
- ↑ Kuhn, R. M. (2009). "The UCSC Genome Browser Database: update 2009". Nucleic Acids Research. 37 (Database): D755–D761. doi:10.1093/nar/gkn875. PMC 2686463
 . PMID 18996895.
. PMID 18996895. - ↑ "WS202 Release Letter". WormBase. 29 May 2009. Retrieved 2013-11-19.
- ↑ "WS197 Release Letter". WormBase. 27 November 2008. Retrieved 2013-11-19.
- ↑ "Genome sequence changes". WormBase. 15 June 2011. Retrieved 2011-08-13.
- ↑ Fire, A; Xu, S; Montgomery, MK; Kostas, SA; Driver, SE; Mello, CC (1998). "Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans". Nature. 391 (6669): 806–11. Bibcode:1998Natur.391..806F. doi:10.1038/35888. PMID 9486653.
- ↑ Harris, TW; et al. (2009-11-12). "WormBase: a comprehensive resource for nematode research". Nucleic Acids Res. 38 (Database issue): D463–7. doi:10.1093/nar/gkp952. PMC 2808986
 . PMID 19910365. Retrieved 2010-04-26.
. PMID 19910365. Retrieved 2010-04-26.
Further reading
- Bird, J; Bird, AC (1991). The structure of nematodes. Academic Press. pp. 1, 69–70, 152–153, 165, 224–225. ISBN 0-12-099651-0.
- Hope, IA (1999). C. elegans: a practical approach. Oxford University Press. pp. 1–6. ISBN 0-19-963738-5.
- Riddle, DL; Blumenthal, T; Meyer, RJ; Priess, JR (1997). C. elegans II. Cold Spring Harbor Laboratory Press. pp. 1–4, 679–683. ISBN 0-87969-532-3.
External links
| Wikimedia Commons has media related to: |
| Wikispecies has information related to: Caenorhabditis elegans |
- Brenner S (2002) Nature's Gift to Science. In. http://nobelprize.org/nobel_prizes/medicine/laureates/2002/brenner-lecture.pdf (also Horvitz and Sulston lectures)
- WormBase – an extensive online database covering the biology and genomics of C. elegans and other nematodes
- WormAtlas – online database on all aspects of C. elegans anatomy with detailed explanations and high-quality images
- WormBook – online review of C. elegans biology
- AceView WormGenes – another genome database for C. elegans, maintained at the NCBI
- C. elegans II – a free online textbook.
- WormWeb Neural Network – an online tool for visualizing and navigating the connectome of C. elegans
- C. elegans movies – a visual introduction to C. elegans
- View the ce11 genome assembly in the UCSC Genome Browser.