Bacterial taxonomy

Bacterial taxonomy is the taxonomy, i.e. the rank-based classification, of bacteria.
In the scientific classification established by Carl von Linné,[1] each species has to be assigned to a genus (binary nomenclature), which in turn is a lower level of a hierarchy of ranks (family, suborder, order, subclass, class, division/phyla, kingdom and domain). In the currently accepted classification of life, there are three domains (Eukaryotes, Bacteria and Archaea),[2] which, in terms of taxonomy, despite following the same principles have several different conventions between them and between their subdivisions as are studied by different disciplines (botany, zoology, mycology and microbiology), for example in zoology there are type specimens, whereas in microbiology there are type strains.
Diversity
Prokaryotes share many common features, such as lack of nuclear membrane, unicellularity, division by binary-fission and generally small size. The various species differ amongst each other based on several characteristics, allowing their identification and classification. Examples include:
- Phylogeny: All bacteria stem from a common ancestor and diversified since, consequently possess different levels of evolutionary relatedness (see Bacterial phyla and Timeline of evolution)
- Metabolism: Different bacteria may have different metabolic abilities (see Microbial metabolism)
- Environment: Different bacteria thrive in different environments, such as high/low temperature and salt (see Extremophiles)
- Morphology: There are many structural differences between bacteria, such as cell shape, Gram stain (number of lipid bilayers) or bilayer composition (see Bacterial cellular morphologies, Bacterial cell structure)
- Pathogenicity: Some bacteria are pathogenic to plants or animals (see Pathogenic bacteria)
Classification history
Bacteria were first observed by Antonie van Leeuwenhoek in 1676, using a single-lens microscope of his own design.[3] He called them "animalcules" and published his observations in a series of letters to the Royal Society.[4][5][6] O. F. Müller (1773, 1786) described eight species of the genus Vibrio (in Infusoria), three of which were spirilliforms.[7] The term Bacterium (a genus) was introduced much later, by Christian Gottfried Ehrenberg in 1838.[8]
Classical classification
Placement

Bacteria were first classified as plants constituting the class Schizomycetes, which along with the Schizophyceae (blue green algae/Cyanobacteria) formed the phylum Schizophyta.[10]
Haeckel in 1866 placed the group in the phylum Moneres (from μονήρης: simple) in the kingdom Protista and defines them as completely structureless and homogeneous organisms, consisting only of a piece of plasma.[9] He subdivided the phylum into two groups:[9]
- die Gymnomoneren (no envelope)
- Protogenes—such as Protogenes primordialis, now classed as a eukaryote and not a bacterium
- Protamaeba—now classed as a eukaryote and not a bacterium
- Vibrio—a genus of comma shaped bacteria first described in 1854[11])
- Bacterium—a genus of rod shaped bacteria first described in 1828, that later gave its name to the members of the Monera, formerly referred to as "a moneron" (plural "monera") in English and "eine Moneren"(fem. plural "Moneres") in German
- Bacillus—a genus of spore-forming rod shaped bacteria first described in 1835[12]
- Spirochaeta—thin spiral shaped bacteria first described in 1835 [13]
- Spirillum—spiral shaped bacteria first described in 1832[14]
- etc.
- die Lepomoneren (with envelope)
- Protomonas—now classed as a eukaryote and not a bacterium. The name was reused in 1984 for an unrelated genus of Bacteria[15]
- Vampyrella—now classed as a eukaryote and not a bacterium
The group was later reclassified as the Prokaryotes by Chatton.[16]
The classification of Cyanobacteria (colloquially "blue green algae") has been fought between being algae or bacteria (for example, Haeckel classified Nostoc in the phylum Archephyta of Algae[9]).
in 1905 Erwin F. Smith accepted 33 valid different names of bacterial genera and over 150 invalid names,[17] and in 1913 Vuillemin[18] in a study concluded that all species of the Bacteria should fall into the genera Planococcus, Streptococcus, Klebsiella, Merista, Planomerista, Neisseria, Sarcina, Planosarcina, Metabacterium, Clostridium, Serratia, Bacterium and Spirillum.
Ferdinand Cohn [19] recognized 4 tribes: Spherobacteria, Microbacteria, Desmobacteria, and Spirobacteria. Stanier and van Neil [20] recognized the Kingdom Monera with 2 phyla, Myxophyta and Schizomycetae, the latter comprising classes Eubacteriae (3 orders), Myxobacteriae (1 order), and Spirochetae (1 order). Bisset [21] distinguished 1 class and 4 orders: Eubacteriales, Actinomycetales, Streptomycetales, and Flexibacteriales. Migula,[22] which was the most widely accepted system of its time and included all then-known species but was based only on morphology, contained the 3 basic groups, Coccaceae, Bacillaceae, and Spirillaceae but also Trichobacterinae for filamentous bacteria; Orla-Jensen [23] established 2 orders: Cephalotrichinae (7 families) and Peritrichinae (presumably with only 1 family). Bergey et al [24] presented a classification which generally followed the 1920 Final Report of the SAB(Society of American Bacteriologists) Committee ( Winslow et al), which divided Class Schizomycetes into 4 orders: Myxobacteriales, Thiobacteriales, Chlamydobacteriales, and Eubacteriales, with a 5th group being 4 genera considered intermediate between bacteria and protozoans: Spirocheta, Cristospira, Saprospira, and Treponema.
However, different authors often reclassified the genera due to the lack of visible traits to go by, resulting in a poor state which was summarised in 1915 by Robert Earle Buchanan [25] By then, the whole group received different ranks and names by different authors namely
- Schizomycetes (Naegeli 1857[10])
- Bacteriaceae (Cohn 1872,[26])
- Bacteria (Cohn 1872b,[27])
- Schizomycetaceae (DeToni and Trevisan 1889,[28])
Furthermore the families into which the class was subdivided, changed from author to author and for some such as Zipf the names where in German and not in Latin [29] The first edition of the Bacteriological Code in 1947 sorted several problems out.[30]
A.R. Prévot's system [31][32]) had 4 subphyla and 8 classes as follows:
Eubacteriales (classes Asporulales and Sporulales) Mycobacteriales (classes Actinomycetales, Myxobacteriales, and Azotobacteriales) Algobacteriales (classes Siderobacteriales and Thiobacteriales) Protozoobacteriales (class Spirochetales)
| Linnaeus 1735[33] |
Haeckel 1866[34] |
Chatton 1925[35] |
Copeland 1938[36] |
Whittaker 1969[37] |
Woese et al. 1990[38] |
Cavalier-Smith 1998[39] |
|---|---|---|---|---|---|---|
| 2 kingdoms | 3 kingdoms | 2 empires | 4 kingdoms | 5 kingdoms | 3 domains | 6 kingdoms |
| (not treated) | Protista | Prokaryota | Monera | Monera | Bacteria | Bacteria |
| Archaea | ||||||
| Eukaryota | Protoctista | Protista | Eucarya | Protozoa | ||
| Chromista | ||||||
| Vegetabilia | Plantae | Plantae | Plantae | Plantae | ||
| Fungi | Fungi | |||||
| Animalia | Animalia | Animalia | Animalia | Animalia |
Subdivisions based on Gram staining
Despite there being little agreement on the major subgroups of the Bacteria, Gram staining results were most commonly used as a classification tool. Consequently, until the advent of molecular phylogeny, the Kingdom Prokaryotae was divided into four divisions,[40] A classification scheme still formally followed by Bergey's manual of systematic bacteriology for tome order[41]
- Gracilicutes (gram-negative)
- Photobacteria (photosynthetic): class Oxyphotobacteriae (water as electron donor, includes the order Cyanobacteriales=blue-green algae, now phylum Cyanobacteria) and class Anoxyphotobacteriae (anaerobic phototrophs, orders: Rhodospirillales and Chlorobiales
- Scotobacteria (non-photosynthetic, now the Proteobacteria and other gram-negative nonphotosynthetic phyla)
- Firmacutes [sic] (gram-positive, subsequently corrected to Firmicutes[42])
- several orders such as Bacillales and Actinomycetales (now in the phylum Actinobacteria)
- Mollicutes (gram variable, e.g. Mycoplasma)
- Mendocutes (uneven gram stain, "methanogenic bacteria", now known as the Archaea)
Molecular era
"Archaic bacteria" and Woese's reclassification
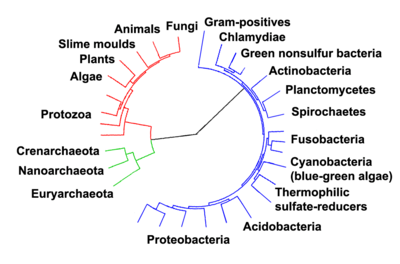
- ^ Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006). "Toward automatic reconstruction of a highly resolved tree of life". Science. 311 (5765): 1283–7. Bibcode:2006Sci...311.1283C. doi:10.1126/science.1123061. PMID 16513982.
Woese argued that the bacteria, archaea, and eukaryotes represent separate lines of descent that diverged early on from an ancestral colony of organisms.[43][44] However, a few biologists argue that the Archaea and Eukaryota arose from a group of bacteria.[45] In any case, it is thought that viruses and archaea began relationships approximately two billion years ago, and that co-evolution may have been occurring between members of these groups.[46] It is possible that the last common ancestor of the bacteria and archaea was a thermophile, which raises the possibility that lower temperatures are "extreme environments" in archaeal terms, and organisms that live in cooler environments appeared only later.[47] Since the Archaea and Bacteria are no more related to each other than they are to eukaryotes, the term prokaryote's only surviving meaning is "not a eukaryote", limiting its value.[48]
With improved methodologies it became clear that the methanogenic bacteria were profoundly different and were (erroneously) believed to be relics of ancient bacteria[49] thus Carl Woese, regarded as the forerunner of the molecular phylogeny revolution, identified three primary lines of descent: the Archaebacteria, the Eubacteria, and the Urkaryotes, the latter now represented by the nucleocytoplasmic component of the Eukaryotes.[50] These lineages were formalised into the rank Domain (regio in Latin) which divided Life into 3 domains: the Eukaryota, the Archaea and the Bacteria.[2] This scheme is still followed today.
Subdivisions
In 1987 Carl Woese divided the Eubacteria into 11 divisions based on 16S ribosomal RNA (SSU) sequences, which with several additions are still used today.[51][52]
Opposition
While the three domain system is widely accepted,[53] some authors have opposed it for various reasons.
One prominent scientist who opposes the three domain system is Thomas Cavalier-Smith, who proposed that the Archaea and the Eukaryotes (the Neomura) stem from Gram positive bacteria (Posibacteria), which in turn derive from gram negative bacteria (Negibacteria) based on several logical arguments,[54][55] which are highly controversial and generally disregarded by the molecular biology community (c.f. reviewers' comments on,[55] e.g. Eric Bapteste is "agnostic" regarding the conclusions) and are often not mentioned in reviews (e.g.[56]) due to the subjective nature of the assumptions made.[57]
However, despite there being a wealth of statistically supported studies towards the rooting of the tree of life between the Bacteria and the Neomura by means of a variety of methods,[58] including some that are impervious to accelerated evolution—which is claimed by Cavalier-Smith to be the source of the supposed fallacy in molecular methods[54]—there are a few studies which have drawn different conclusions, some of which place the root in the phylum Firmicutes with nested archaea.[59][60][61]
Radhey Gupta's molecular taxonomy, based on conserved signature sequences of proteins, includes a monophyletic Gram negative clade, a monophyletic Gram positive clade, and a polyphyletic Archeota derived from Gram positives.[62][63][64] Hori and Osawa's molecular analysis indicated a link between Metabacteria (=Archeota) and eukaryotes.[65] The only cladistic analyses for bacteria based on classical evidence largely corroborate Gupta's results (see comprehensive mega-taxonomy).
James Lake presented a 2 primary kingdom arrangement (Parkaryotae + eukaryotes and eocytes + Karyotae) and suggested a 5 primary kingdom scheme (Eukaryota, Eocyta, Methanobacteria, Halobacteria, and Eubacteria) based on ribosomal structure and a 4 primary kingdom scheme (Eukaryota, Eocyta, Methanobacteria, and Photocyta), bacteria being classified according to 3 major biochemical innovations: photosynthesis (Photocyta), methanogenesis (Methanobacteria), and sulfur respiration (Eocyta).[66][67][68] He has also discovered evidence that Gram-negative bacteria arose from a symbiosis between 2 Gram-positive bacteria.[69]
Authorities
Classification is the grouping of organisms into progressively more inclusive groups based on phylogeny and phenotype, while nomenclature is the application of formal rules for naming organisms.[70]
Nomenclature authority
Despite there being no official and complete classification of prokaryotes, the names (nomenclature) given to prokaryotes are regulated by the International Code of Nomenclature of Bacteria (Bacteriological Code), a book which contains general considerations, principles, rules, and various notes, and advises[71] in a similar fashion to the nomenclature codes of other groups.
Classification authorities
The taxa which have been correctly described are reviewed in Bergey's manual of Systematic Bacteriology, which aims to aid in the identification of species and is considered the highest authority.[41] An online version of the taxonomic outline of bacteria and archaea is available .
LPSN is an online database which currently contains over two thousand accepted names with their references, etymologies and various notes.[72]
Description of new species
The International Journal of Systematic Bacteriology/International Journal of Systematic and Evolutionary Microbiology (IJSB/IJSEM) is a peer reviewed journal which acts as the official international forum for the publication of new prokaryotic taxa. If a species is published in a different peer review journal, the author can submit a request to IJSEM with the appropriate description, which if correct, the new species will be featured in the Validation List of IJSEM.
Distribution
Microbial culture collections are depositories of strains which aim to safeguard them and to distribute them. The main ones being:[70]
| Collection Acronym | Name | Location |
|---|---|---|
| ATCC | American Type Culture Collection | Manassas, Virginia |
| NCTC | National Collection of Type Cultures | Public Health England, United Kingdom |
| BCCM | Belgium Coordinated Collection of Microorganisms | Ghent, Belgium |
| CIP | Collection d'Institut Pasteur | Paris, France |
| DSMZ | Deutsche Sammlung von Mikroorganismen und Zellkulturen | Braunschweig, Germany |
| JCM | Japan Collection of Microorganisms | Saitama, Japan |
| NCCB | Netherlands Culture Collection of Bacteria | Utrecht, Netherlands |
| NCIMB | National Collection of industrial, Marine and food bacteria | Aberdeen, Scotland |
| ICMP | International Collection of Microorganisms from Plants | Auckland, New Zealand |
Analyses
Bacteria were at first classified based solely on their shape (vibrio, bacillus, coccus etc.), presence of endospores, gram stain, aerobic conditions and motility. This system changed with the study of metabolic phenotypes, where metabolic characteristics were used.[73] Recently, with the advent of molecular phylogeny, several genes are used to identify species, the most important of which is the 16S rRNA gene, followed by 23S, ITS region, gyrB and others to confirm a better resolution. The quickest way to identify to match an isolated strain to a species or genus today is done by amplifying it's 16S gene with universal primers and sequence the 1.4kb amplicon and submit it to a specialised web-based identification database, namely either Ribosomal Database Project, which align the sequence to other 16S sequences using infernal, a secondary structure bases global alignment,[74][75] or ARB SILVA, which aligns sequences via SINA (SILVA incremental aligner), which does a local alignment of a seed and extends it .[76]
Several identification methods exists:[70]
- Phenotypic analyses
- fatty acid analyses
- Growth conditions (Agar plate, Biolog multiwell plates)
- Genetic analyses
- DNA-DNA hybridization
- DNA profiling
- Sequence
- GC ratios
- Phylogenetic analyses
- 16S-based phylogeny
- phylogeny based on other genes
- Multi-gene sequence analysis
- Whole-genome sequence based analysis
New species
The minimal standards for describing a new species depend on which group the species belongs to. c.f.[77]
Candidatus
Candidatus is a component of the taxonomic name for a bacterium that cannot be maintained in a Bacteriology Culture Collection. It is an interim taxonomic status for noncultivable organisms. e.g. "Candidatus Pelagibacter ubique"
Species concept
Bacteria divide asexually and for the most part do not show regionalisms ("Everything is everywhere"), therefore the concept of species, which works best for animals, becomes entirely a matter of judgement.
The number of named species of bacteria and archaea (approximately 13,000)[78] is surprisingly small considering their early evolution, genetic diversity and residence in all ecosystems. The reason for this is the differences in species concepts between the bacteria and macro-organisms, the difficulties in growing/characterising in pure culture (a prerequisite to naming new species, vide supra) and extensive horizontal gene transfer blurring the distinction of species.[79]
The most commonly accepted definition is the polyphasic species definition, which takes into account both phenotypic and genetic differences.[80] However, a quicker diagnostic ad hoc threshold to separate species is less than 70% DNA–DNA hybridisation,[81] which corresponds to less than 97% 16S DNA sequence identity.[82] It has been noted that if this were applied to animal classification, the order primates would be a single species.[83]
Pathology vs. phylogeny
Ideally, taxonomic classification should reflect the evolutionary history of the taxa, i.e. the phylogeny. Although some exceptions are present when the phenotype differs amongst the group, especially from a medical standpoint. Some examples of problematic classifications follow.
Escherichia coli: overly large and polyphyletic
In the Enterobacteriaceae family of the class Gammaproteobacteria, the species in the genus Shigella (S. dysenteriae, S. flexneri, S. boydii, S. sonnei) by an evolutionary point of view are strains of the species Escherichia coli (polyphyletic), but due to genetic differences cause different medical conditions in the case of the pathogenic strains.,[84] Escherichia coli is a badly classified species as some strains share only 20% of their genome. Being so diverse it should be given a higher taxonomic ranking. However, due to the medical conditions associated with the species, it will not be changed to avoid confusion in medical context.
Bacillus cereus group: close and polyphyletic
In a similar way, the Bacillus species (=phylum Firmicutes) belonging to the "B. cereus group" (B. anthracis, B. cereus, B . thuringiensis, B. mycoides, B. pseudomycoides, B. weihenstephanensis and B. medusa) have 99-100% similar 16S rRNA sequence (97% is a commonly cited adequate species cut-off) and are polyphyletic, but for medical reasons (anthrax etc.) remain separate.[85]
Yersinia pestis: extremely recent species
Yersinia pestis is in effect a strain of Yersinia pseudotuberculosis, but with a pathogenicity island that confer a drastically different pathology (Black plague and tuberculosis-like symptoms respectively) which arose 15,000 to 20,000 years ago.[86]
Nested genera in Pseudomonas
In the gammaproteobacterial order Pseudomonadales, the genus Azotobacter and the species Azomonas macrocytogenes are actually members of the genus Pseudomonas, but were misclassified due to nitrogen fixing capabilities and the large size of the genus Pseudomonas which renders classification problematic.[73][87][88] This will probably rectified in the close future.
Nested genera in Bacillus
Another example of a large genus with nested genera is the Bacillus genus, in which the genera Paenibacillus and Brevibacillus are nested clades.[89] There is insufficient genomic data at present to fully and effectively correct taxonomic errors in Bacillus.
Agrobacterium: resistance to name change
Based on molecular data it was shown that the genus Agrobacterium is nested in Rhizobium and the Agrobacterium species transferred to the Rhizobium genus (resulting in the following comp. nov.: Rhizobium radiobacter (formerly known as A. tumefaciens), R. rhizogenes, R. rubi, R. undicola and R. vitis)[90] Given the plant pathogenic nature of Agrobacterium species, it was proposed to maintain the genus Agrobacterium[91] and the latter was counter-argued[92]
Nomenclature
Taxonomic names are written in italics (or underlined when handwritten) with a majuscule first letter with the exception of epithets for species and subspecies. Despite it being common in zoology, tautonyms (e.g. Bison bison) are not acceptable and names of taxa used in zoology, botany or mycology cannot be reused for Bacteria (Botany and Zoology do share names).
Nomenclature is the set of rules and conventions which govern the names of taxa. The difference in nomenclature between the various kingdoms/domains is reviewed in.[93]
For Bacteria, valid names must have a Latin or Neolatin name and can only use basic latin letters (w and j inclusive, see History of the Latin alphabet for these), consequently hyphens, accents and other letters are not accepted and should be transliterated correctly (e.g. ß=ss).[94] Ancient Greek being written in the Greek alphabet, needs to be transliterated into the Latin alphabet.
When compound words are created, a connecting vowel is needed depending on the origin of the preceding word, regardless of the word that follows, unless the latter starts with a vowel in which case no connecting vowel is added. If the first compound is Latin then the connecting vowel is an -i-, whereas if the first compound is Greek, the connecting vowel is an -o-.[95]
For etymologies of names consult LPSN.
Rules for higher taxa
For the Prokaryotes (Bacteria and Archaea) the rank kingdom is not used[96] (although some authors refer to phyla as kingdoms[70])
If a new or amended species is placed in new ranks, according to Rule 9 of the Bacteriologocal Code the name is formed by the addition of an appropriate suffix to the stem of the name of the type genus.[71] For subclass and class the recommendation from [97] is generally followed, resulting in a neutral plural, however a few names do not follow this and instead keep into account graeco-latin grammar (e.g. the female plurals Thermotogae, Aquificae and Chlamydiae, the male plurals Chloroflexi, Bacilli and Deinococci and the greek plurals Spirochaetes, Gemmatimonadetes and Chrysiogenetes).[98]
| Rank | Suffix | Example |
|---|---|---|
| Genus | Elusimicrobium | |
| Subtribe (disused) | -inae | (Elusimicrobiinae) |
| Tribe (disused) | -inae | (Elusimicrobiieae) |
| Subfamily | -oideae | (Elusimicrobioideae) |
| Family | -aceae | Elusimicrobiaceae |
| Suborder | -ineae | (Elusimicrobineae) |
| Order | -ales | Elusimicrobiales |
| Subclass | -idae | (Elusimicrobidae) |
| Class | -ia | Elusimicrobia |
| Phylum | see text | Elusimicrobia |
Phyla endings
Phyla are not covered by the Bacteriological code,[98] however, the scientific community generally follows the Ncbi and Lpsn taxonomy, where the name of the phylum is generally the plural of the type genus, with the exception of the Firmicutes, Cyanobacteria and Proteobacteria, whose names do not stem from a genus name. The higher taxa proposed by Cavalier-Smith[54] are generally disregarded by the molecular phylogeny community (e.g.[56]) (vide supra).
For the Archaea the suffix -archaeota is used.[99] For bacterial phyla it was proposed that the suffix -bacteria be used for phyla.[100]
Consequently for main phyla the name of the phyla is the same as the first described class:
- Acidobacteria (from Acidobacterium)
- Actinobacteria (from Actinomyces)
- Caldisericia (from Caldisericum)
- Elusimicrobia (from Elusimicrobium)
- Fusobacteria (from Fusobacterium)
- Thermodesulfobacteria (from Thermodesulfobacterium)
- Thermotogae (from Thermotoga)
- Aquificae (from Aquifex)
- Chlamydiae (from Chlamydia)
- Chloroflexi (from Chloroflexus)
- Chrysiogenetes (from Chrysiogenes)
- Gemmatimonadetes (from Gemmatimonas)
- Deferribacteres (from Deferribacter)
Whereas for others where the -ia suffix for class is used regardless of grammar they differ:
- phylum Bacteroidetes vs. class Bacteroidia from Bacteroides
- phylum Chlorobi vs. class Chlorobia from Chlorobium
- phylum Verrucomicrobia vs. class Verrucomicrobiae from Verrucomicrobium (anomalous class name)
- phylum Dictyoglomi versus class Dictyoglomia from Dictyoglomus
- phylum Fibrobacteres versus class Fibrobacteria from Fibrobacter (c.f. the suffix -bacter, note the difference with Deferribacteres)
- phylum Lentisphaerae versus class Lentisphaeria from Lentisphaera
- phylum Nitrospira or Nitrospirae versus class Nitrospira from Nitrospira
- phylum Spirochaetes versus class Spirochaetae from Spirochaeta
- phylum Synergistetes versus class Synergistetia from Synergistes
- phylum Planctomycetes versus Planctomycea from Planctomyces
An exception is the phylum Deinococcus-Thermus, which bears a hyphenated pair of genera —only non accented latin letters are accepted for valid names, but phyla are not officially recognised.[99]
Names after people
Several species are named after people, either the discoverer or a famous person in the field of microbiology, for example Salmonella is after D.E. Salmon, who discovered it (albeit as "Bacillus typhi"[101]).[102]
For the generic epithet, all names derived from people must be in the female nominative case, either by changing the ending to -a or to the diminutive -ella, depending on the name.[95]
For the specific epithet, the names can be converted into either adjectival form (adding -nus (m.), -na (f.), -num (n.) according to the gender of the genus name) or the genitive of the latinised name.[95]
Names after places
Many species (the specific epithet) are named after the place they are present or found (e.g. Thiospirillum jenense). Their names are created by forming an adjective by joining the locality's name with the ending -ensis (m. or f.) or ense (n.) in agreement with the gender of the genus name, unless a classical Latin adjective exists for the place. However, names of places should not be used as nouns in the genitive case.[95]
Vernacular names
Despite the fact that some hetero/homogeneus colonies or biofilms of bacteria have names in English (e.g. dental plaque or Star jelly), no bacterial species has a vernacular/trivial/common name in English.
For names in the singular form, plurals cannot be made (singulare tantum) as would imply multiple groups with the same label and not multiple members of that group (by analogy, in English, chairs and tables are types of furniture, which cannot be used in the plural form "furnitures" to describe both members), conversely names plural form are pluralia tantum. However, a partial exception to this is made by the use of vernacular names. However, to avoid repetition of taxonomic names which break the flow of prose, vernacular names of members of a genus or higher taxa are often used and recommended, these are formed by writing the name of the taxa in sentence case roman ("standard" in MS Office) type, therefore treating the proper noun as an English common noun (e.g. the salmonellas), although there is some debate about the grammar of plurals, which can either be regular plural by adding -(e)s (the salmonellas) or using the ancient Greek or Latin plural form (irregular plurals) of the noun (the salmonellae); the latter is problematic as the plural of - bacter would be -bacteres, while the plural of myces (N.L. masc. n. from Gr. masc. n. mukes) is mycetes.[103]
Customs are present for centain names, such as those ending in -monas are converted into -monad (one pseudomonad, two aeromonads and not -monades).
Bacteria which are the etiological cause for a disease are often referred to by the disease name followed by a describing noun (bacterium, bacillus, coccus, agent or the name of their phylum) e.g. cholera bacterium (Vibrio cholerae) or Lyme disease spirochete (Borrelia burgdorferi) , note also rickettsialpox (Rickettsia akari) (for more see [104]).
Treponema is converted into treponeme and the plural is treponemes and not treponemata.
Some unusual bacteria have special names such as Quin's oval (Quinella ovalis) and Walsby's square (Haloquadratum walsbyi).
Before the advent of molecular phylogeny, many higher taxonomic groupings had only trivial names, which are still used today, some of which are polyphyletic, such as Rhizobacteria. Some higher taxonomic trivial names are:
- Blue-green algae are members of the phylum Cyanobacteria
- Green non-sulfur bacteria are members of the phylum Chloroflexi
- Green sulfur bacteria are members of the Chlorobi
- Purple bacteria are some, but not all, members of the phylum Proteobacteria
- Purple sulfur bacteria are members of the order Chromatiales
- low G+C Gram-positive bacteria are members of the phylum Firmicutes, regardless of GC content
- high G+C Gram-positive bacteria are members of the phylum Actinobacteria, regardless of GC content
- Rhizobacteria are members of various genera of proteobacteria
- Rhizobia are members of the order Rhizobiales
- Lactic streptococci are members of the genus Lactococcus
- Coryneform bacteria are members of the family Corynebacteriaceae
- Fruiting gliding bacteria or myxobacteria are members of the order Myxococcales
- Enterics are members of the order Enterobacteriales, although the term is avoided if they do not live in the intestines, such as Pectobacterium
- Acetic acid bacteria are members of the family Acetobacteraceae
Terminology
- The abbreviation for species is sp. (plural spp.) and is used after a generic epithet to indicate a species of that genus. Often used to denote a strain of a genus for which the species is not known either because has the organism has not been described yet as a species or insufficient tests were conducted to identify it. For example Halomonas sp. GFAJ-1
- If a bacterium is known and well-studied but not culturable, it is given the term Candidatus in its name
- A basonym is original name of a new combination, namely the first name given to a taxa before it was reclassified
- A synonym is an alternative name for a taxa, i.e. a taxa was erroneusly described twice
- When a taxon is transferred it becomes a new combination (comb. nov.) or nomina nova (nom. nov.)
- paraphyly, monophyly and polyphyly
See also
- List of Latin and Greek words commonly used in systematic names
- List of Bacteria genera
- List of bacterial orders
- List of sequenced prokaryotic genomes
- Human microbiome project
- Microbial ecology
- List of clinically important bacteria
- List of sequenced archaeal genomes
- List of Archaea genera
- Species problem
- Evolutionary grade
- Cryptic species complex
- Synonym (taxonomy)
- Taxonomy
- LPSN, list of accepted bacterial and archaeal names
- Cyanobacteria, a phyla of common bacteria but poor classified at present
- Bacterial phyla, a complicated classification
References
- ↑ Linnaeus, Carl (1735). Systemae Naturae, sive regna tria naturae, systematics proposita per classes, ordines, genera & species.
- 1 2 Woese, C. R.; Kandler, O.; Wheelis, M. L. (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences. 87 (12): 4576–4579. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159
 . PMID 2112744.
. PMID 2112744. - ↑ Porter JR (1976). "Antony van Leeuwenhoek: tercentenary of his discovery of bacteria". Bacteriological Reviews. 40 (2): 260–9. PMC 413956
 . PMID 786250.
. PMID 786250. - ↑ van Leeuwenhoek A (1684). "An abstract of a letter from Mr. Anthony Leevvenhoek at Delft, dated Sep. 17, 1683, Containing Some Microscopical Observations, about Animals in the Scurf of the Teeth, the Substance Call'd Worms in the Nose, the Cuticula Consisting of Scales". Philosophical Transactions (1683–1775). 14 (155–166): 568–574. doi:10.1098/rstl.1684.0030. Retrieved 2007-08-19.
- ↑ van Leeuwenhoek A (1700). "Part of a Letter from Mr Antony van Leeuwenhoek, concerning the Worms in Sheeps Livers, Gnats, and Animalcula in the Excrements of Frogs". Philosophical Transactions (1683–1775). 22 (260–276): 509–518. doi:10.1098/rstl.1700.0013. Retrieved 2007-08-19.
- ↑ van Leeuwenhoek A (1702). "Part of a Letter from Mr Antony van Leeuwenhoek, F. R. S. concerning Green Weeds Growing in Water, and Some Animalcula Found about Them". Philosophical Transactions (1683–1775). 23 (277–288): 1304–11. doi:10.1098/rstl.1702.0042. Retrieved 2007-08-19.
- ↑ Pot, B., Gillis, M., and De Ley, J., The genus Aquaspirillum. In: Balows, A., Trüper, H.G., Dworkin, M., Harder, W., Schleifer, K.-H. (Eds.). The prokaryotes, 2nd ed, vol. 3. Springer-Verlag. New York. 1991
- ↑ "Etymology of the word "bacteria"". Online Etymology dictionary. Archived from the original on 18 November 2006. Retrieved 2006-11-23.
- 1 2 3 4 Haeckel, Ernst (1867). Generelle Morphologie der Organismen. Reimer, Berlin. ISBN 1-144-00186-2.
- 1 2 C. Von Nägeli (1857). R. Caspary, ed. "Bericht über die Verhandlungen der 33. Versammlung deutscher Naturforscher und Aerzte, gehalten in Bonn von 18 bis 24 September 1857" [Report on the negotiations on 33 Meeting of German Natural Scientists and Physicians, held in Bonn, 18 to 24 September 1857]. Botanische Zeitung. 15: 749–776. <https://archive.org/stream/botanischezeitun15mohl#page/372/mode/2up>
- ↑ PACINI (F.): Osservazione microscopiche e deduzioni patologiche sul cholera asiatico. Gazette Medicale de Italiana Toscano Firenze, 1854, 6, 405-412.
- ↑ EHRENBERG (C.G.): Dritter Beitrag zur Erkenntniss grosser Organisation in der Richtung des kleinsten Raumes. Physikalische Abhandlungen der Koeniglichen Akademie der Wissenschaften zu Berlin aus den Jahren 1833-1835, 1835, pp. 143-336.
- ↑ EHRENBERG (C.G.): Dritter Beitrag zur Erkenntniss grosser Organisation in der Richtung des kleinsten Raumes. Abhandlungen der Preussischen Akademie der Wissenschaften (Berlin) aus den Jahre 1833-1835, pp. 143-336.
- ↑ EHRENBERG (C.G.): Beiträge zur Kenntnis der Organization der Infusorien und ihrer geographischen Verbreitung besonders in Sibirien. Abhandlungen der Koniglichen Akademie der Wissenschaften zu Berlin, 1832, 1830, 1-88.
- ↑ Protomonas entry in LPSN [Euzéby, J.P. (1997). "List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet". Int J Syst Bacteriol. 47 (2): 590–2. doi:10.1099/00207713-47-2-590. ISSN 0020-7713. PMID 9103655.]
- ↑ É. Chatton (1925). "Pansporella perplexa. Réflexions sur la biologie et la phylogénie des protozoaires". Ann. Sci. Nat. Zool. 10-VII: 1–84.
- ↑ Erwin F. Smith Nomenclature and Classification in Bacteria in Relation to Plant Diseases vol. 1 (1905)
- ↑ Vuillemin. 1913. Genera Schizomycetum. Annales Mycologici. 11,512-527.
- ↑ Cohn, Ferdinand 1875, Untersuchungen uber Bakterien, II. Beitrage zur Biologie der Pfanzen 1: 141-207
- ↑ Stanier and van Neil,1941, The main outlines of bacterial classification. J Bacteriol 42: 437- 466
- ↑ Bisset, 1962, Bacteria, 2nd ed., Livingston, London
- ↑ 1897, System der Bakterien. Gustav Fischer, Jena
- ↑ Orla-Jensen, 1909, Die Hauptlinien des naturalischen Bakteriensystems nebst einer Ubersicht der Garungsphenomene. Zentr. Bakt. Parasitenk., II, 22: 305-346
- ↑ 1925, Bergey's Manual of Determinative Bacteriology, Baltimore : Williams & Wilkins Co. ( with many subsequent editions)
- ↑ Buchanan, R E J Bacteriol. 1916 Nov;1(6):591-6.ISSN 0021-9193 PMCID 378679
- ↑ COHN, FERDINAND 1872 a Organismen in der Pockenlymphe. Virchow's Archiv, 55, 229-238. p. 237
- ↑ COHN, FERDINAND 1872 b Untersuchungen ilber Bakterien. Beitrage zur Biologie der Pflanzen, 1 (Heft 1), 127-224. p. 136
- ↑ DETONI, J. B., AND TREVISAN, V. 1889 Schizomycetaceae, in Saccardo's Sylloge Fungorum, 8, 923-1087. p. 923
- ↑ Buchanan RE (March 1917). "Studies in the Nomenclature and Classification of the Bacteria: II. The Primary Subdivisions of the Schizomycetes". J. Bacteriol. 2 (2): 155–64. PMC 378699
 . PMID 16558735.
. PMID 16558735. - ↑ "International Bacteriological Code of Nomenclature". J. Bacteriol. 55 (3): 287–306. March 1948. PMC 518444
 . PMID 16561459.
. PMID 16561459. - ↑ Prévot, A.R. 1958. Manuel de classification et détermination des bactéries anaérobies. Masson, Paris
- ↑ Prévot, A.R. Bactéries in Précis de Botanique, 1963, Masson, Paris
- ↑ Linnaeus, C. (1735). Systemae Naturae, sive regna tria naturae, systematics proposita per classes, ordines, genera & species.
- ↑ Haeckel, E. (1866). Generelle Morphologie der Organismen. Reimer, Berlin.
- ↑ Chatton, É. (1925). "Pansporella perplexa. Réflexions sur la biologie et la phylogénie des protozoaires". Annales des Sciences Naturelles - Zoologie et Biologie Animale. 10-VII: 1–84.
- ↑ Copeland, H. (1938). "The kingdoms of organisms". Quarterly Review of Biology. 13: 383–420. doi:10.1086/394568.
- ↑ Whittaker, R. H. (January 1969). "New concepts of kingdoms of organisms". Science. 163 (3863): 150–60. Bibcode:1969Sci...163..150W. doi:10.1126/science.163.3863.150. PMID 5762760.
- ↑ Woese, C.; Kandler, O.; Wheelis, M. (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya.". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–9. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159
 . PMID 2112744.
. PMID 2112744. - ↑ Cavalier-Smith, T. (1998). "A revised six-kingdom system of life". Biological Reviews. 73 (03): 203–66. doi:10.1111/j.1469-185X.1998.tb00030.x. PMID 9809012.
- ↑ GIBBONS N.E.; MURRAY R.G.E. (1978). "Proposals concerning the higher taxa of bacteria". International Journal of Systematic Bacteriology. 28: 1–6. doi:10.1099/00207713-28-1-1.
- 1 2 George M. Garrity: Bergey's manual of systematic bacteriology. 2. Auflage. Springer, New York, 2005, Volume 2: The Proteobacteria, Part B: The Gammaproteobacteria
- ↑ MURRAY (R.G.E.): The higher taxa, or, a place for everything...? In: N.R. KRIEG and J.G. HOLT (ed.) Bergey's Manual of Systematic Bacteriology, vol. 1, The Williams & Wilkins Co., Baltimore, 1984, p. 31-34
- ↑ Woese CR, Gupta R (1981). "Are archaebacteria merely derived 'prokaryotes'?". Nature. 289 (5793): 95–6. Bibcode:1981Natur.289...95W. doi:10.1038/289095a0. PMID 6161309.
- ↑ Woese C (1998). "The universal ancestor". Proceedings of the National Academy of Sciences of the United States of America. 95 (12): 6854–9. Bibcode:1998PNAS...95.6854W. doi:10.1073/pnas.95.12.6854. PMC 22660
 . PMID 9618502.
. PMID 9618502. - ↑ Gupta RS (2000). "The natural evolutionary relationships among prokaryotes". Crit. Rev. Microbiol. 26 (2): 111–31. doi:10.1080/10408410091154219. PMID 10890353.
- ↑ C. Michael Hogan. 2010. Virus. Encyclopedia of Earth. Editors: Cutler Cleveland and Sidney Draggan
- ↑ Gribaldo S, Brochier-Armanet C (2006). "The origin and evolution of Archaea: a state of the art". Philosophical Transactions of the Royal Society B. 361 (1470): 1007–22. doi:10.1098/rstb.2006.1841. PMC 1578729
 . PMID 16754611.
. PMID 16754611. - ↑ Woese CR (1 March 1994). "There must be a prokaryote somewhere: microbiology's search for itself". Microbiol. Rev. 58 (1): 1–9. PMC 372949
 . PMID 8177167.
. PMID 8177167. - ↑ Balch, W.; Magrum, L.; Fox, G.; Wolfe, R.; Woese, C. (1977). "An ancient divergence among the bacteria". Journal of Molecular Evolution. 9 (4): 305–311. doi:10.1007/BF01796092. PMID 408502.
- ↑ Francolini, C.; Egeth, H. (1979). "Perceptual selectivity is task dependent: The pop-out effect poops out". Perception & psychophysics. 25 (2): 99–110. doi:10.3758/bf03198793. PMID 432104.
- ↑ Holland L. (22 May 1990). "Woese,Carl in the forefront of bacterial evolution revolution". Scientist. 4 (10).
- ↑ Woese, C. R. (1987). "Bacterial evolution". Microbiological reviews. 51 (2): 221–271. PMC 373105
 . PMID 2439888.
. PMID 2439888. - ↑ Ward, B. B. (2002). "How many species of prokaryotes are there?". Proceedings of the National Academy of Sciences. 99 (16): 10234–10236. Bibcode:2002PNAS...9910234W. doi:10.1073/pnas.162359199.
- 1 2 3 Cavalier-Smith, T (2002). "The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification". International Journal of Systematic and Evolutionary Microbiology. 52 (Pt 1): 7–76. doi:10.1099/00207713-52-1-7. PMID 11837318.
- 1 2 Cavalier-Smith, T. (2006). "Rooting the tree of life by transition analyses". Biology Direct. 1: 19. doi:10.1186/1745-6150-1-19. PMC 1586193
 . PMID 16834776.
. PMID 16834776. - 1 2 Pace, N. R. (2009). "Mapping the Tree of Life: Progress and Prospects". Microbiology and Molecular Biology Reviews. 73 (4): 565–576. doi:10.1128/MMBR.00033-09. PMC 2786576
 . PMID 19946133.
. PMID 19946133. - ↑ Fournier, G. P.; Gogarten, J. P. (2010). "Rooting the Ribosomal Tree of Life". Molecular Biology and Evolution. 27 (8): 1792–1801. doi:10.1093/molbev/msq057. PMID 20194428.
- ↑ Fournier, G. P.; Gogarten, J. P. (2010). "Rooting the Ribosomal Tree of Life". Molecular Biology and Evolution. 27 (8): 1792–1801. doi:10.1093/molbev/msq057. PMID 20194428.
- ↑ Lake, J.; Servin, J.; Herbold, C.; Skophammer, R. (2008). "Evidence for a New Root of the Tree of Life". Systematic Biology. 57 (6): 835–843. doi:10.1080/10635150802555933. PMID 19085327.
- ↑ Skophammer, R. G.; Servin, J. A.; Herbold, C. W.; Lake, J. A. (2007). "Evidence for a Gram-positive, Eubacterial Root of the Tree of Life". Molecular Biology and Evolution. 24 (8): 1761–1768. doi:10.1093/molbev/msm096. PMID 17513883.
- ↑ Valas, R. E.; Bourne, P. E. (2011). "The origin of a derived superkingdom: How a gram-positive bacterium crossed the desert to become an archaeon". Biology Direct. 6: 16. doi:10.1186/1745-6150-6-16. PMC 3056875
 . PMID 21356104.
. PMID 21356104. - ↑ Gupta, R. S. (1998). "Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes". Microbiology and molecular biology reviews : MMBR. 62 (4): 1435–1491. PMC 98952
 . PMID 9841678.
. PMID 9841678. - ↑ Gupta, R. S. (1998). "Life's third domain (Archaea): An established fact or an endangered paradigm?". Theoretical population biology. 54 (2): 91–104. doi:10.1006/tpbi.1998.1376. PMID 9733652.
- ↑ Gupta, R. S. (2000). "The Natural Evolutionary Relationships among Prokaryotes". Critical Reviews in Microbiology. 26 (2): 111–131. doi:10.1080/10408410091154219. PMID 10890353.
- ↑ Hori, H.; Osawa, S. (1987). "Origin and evolution of organisms as deduced from 5S ribosomal RNA sequences". Molecular Biology and Evolution. 4 (5): 445–472. PMID 2452957.
- ↑ Lake, J. A. (1986). "An alternative to archaebacterial dogma". Nature. 319 (6055): 626. Bibcode:1986Natur.319..626L. doi:10.1038/319626b0.
- ↑ Lake, JA; Henderson, E; Oakes, M; Clark, MW (1984). "Eocytes: A new ribosome structure indicates a kingdom with a close relationship to eukaryotes". Proceedings of the National Academy of Sciences of the United States of America. 81 (12): 3786–3790. Bibcode:1984PNAS...81.3786L. doi:10.1073/pnas.81.12.3786. PMC 345305
 . PMID 6587394.
. PMID 6587394. - ↑ Lake, J. A.; Henderson, E.; Clark, M. W.; Scheinman, A.; Oakes, M. I. (1986). "Mapping evolution with three dimensional ribosome structure". Systematic and Applied Microbiology. 7: 131–136. doi:10.1016/S0723-2020(86)80135-7.
- ↑ Lake, J. (2009). "Evidence for an early prokaryotic endosymbiosis". Nature. 460 (7258): 967–971. Bibcode:2009Natur.460..967L. doi:10.1038/nature08183. PMID 19693078.
- 1 2 3 4 Madigan, Michael (2009). Brock Biology of Microorganisms. San Francisco: Pearson/Benjamin Cummings. ISBN 0-13-232460-1.
- 1 2 Lapage, S.; Sneath, P.; Lessel, E.; Skerman, V.; Seeliger, H.; Clark, W. (1992). International Code of Nomenclature of Bacteria: Bacteriological Code, 1990 Revision. Washington, D.C.: ASM Press. PMID 21089234.
- ↑ Introductory entry in LPSN [Euzéby, J.P. (1997). "List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet". Int J Syst Bacteriol. 47 (2): 590–2. doi:10.1099/00207713-47-2-590. ISSN 0020-7713. PMID 9103655.]
- 1 2 Palleroni, N. J. (2010). "The Pseudomonas Story". Environmental Microbiology. 12 (6): 1377–1383. doi:10.1111/j.1462-2920.2009.02041.x. PMID 20553550.
- ↑ Olsen, G.; Larsen, N.; Woese, C. (1991). "The ribosomal RNA database project". Nucleic Acids Research. 19 Suppl: 2017–2021. doi:10.1093/nar/19.suppl.2017. PMC 331344
 . PMID 2041798.
. PMID 2041798. - ↑ Cole, J. R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R. J.; Kulam-Syed-Mohideen, A. S.; McGarrell, D. M.; Marsh, T.; Garrity, G. M.; Tiedje, J. M. (2009). "The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis". Nucleic Acids Research. 37 (Database issue): D141–D145. doi:10.1093/nar/gkn879. PMC 2686447
 . PMID 19004872.
. PMID 19004872. - ↑ Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B. M.; Ludwig, W.; Peplies, J.; Glockner, F. O. (2007). "SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB". Nucleic Acids Research. 35 (21): 7188–7196. doi:10.1093/nar/gkm864. PMC 2175337
 . PMID 17947321.
. PMID 17947321. - ↑ minimal standards entry in LPSN [Euzéby, J.P. (1997). "List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet". Int J Syst Bacteriol. 47 (2): 590–2. doi:10.1099/00207713-47-2-590. ISSN 0020-7713. PMID 9103655.]
- ↑ http://www.bacterio.net/-number.html
- ↑ Staley, J. T. (2006). "The bacterial species dilemma and the genomic-phylogenetic species concept". Philosophical Transactions of the Royal Society B: Biological Sciences. 361 (1475): 1899–1909. doi:10.1098/rstb.2006.1914. PMC 1857736
 . PMID 17062409.
. PMID 17062409. - ↑ Gevers, D.; Cohan, F. M.; Lawrence, J. G.; Spratt, B. G.; Coenye, T.; Feil, E. J.; Stackebrandt, E.; Van De Peer, Y. V.; Vandamme, P.; Thompson, F. L.; Swings, J. (2005). "Opinion: Re-evaluating prokaryotic species". Nature Reviews Microbiology. 3 (9): 733–739. doi:10.1038/nrmicro1236. PMID 16138101.
- ↑ Wayne, L. (1988). "International Committee on Systematic Bacteriology: Announcement of the report of the ad hoc Committee on Reconciliation of Approaches to Bacterial Systematics". Zentralblatt für Bakteriologie, Mikrobiologie, und Hygiene. Series A, Medical microbiology, infectious diseases, virology, parasitology. 268 (4): 433–434. doi:10.1016/s0176-6724(88)80120-2. PMID 3213314.
- ↑ Stackebrandt, E; Goebel, B.M. (1994). "Taxonomic note: a place for DNA–DNA reassociation and 16S rDNA sequence analysis in the present species definition in bacteriology". Int. J. Syst. Bacteriol. 44: 846–849. doi:10.1099/00207713-44-4-846.
- ↑ Staley, J. (1997). "Biodiversity: Are microbial species threatened?". Current opinion in biotechnology. 8 (3): 340–345. doi:10.1016/s0958-1669(97)80014-6. PMID 9206017.
- ↑ Lan, R; Reeves, PR (2002). "Escherichia coli in disguise: molecular origins of Shigella". Microbes and infection / Institut Pasteur. 4 (11): 1125–32. doi:10.1016/S1286-4579(02)01637-4. PMID 12361912.
- ↑ Bavykin, S. G.; Lysov, Y. P.; Zakhariev, V.; Kelly, J. J.; Jackman, J.; Stahl, D. A.; Cherni, A. (2004). "Use of 16S rRNA, 23S rRNA, and gyrB Gene Sequence Analysis to Determine Phylogenetic Relationships of Bacillus cereus Group Microorganisms". Journal of Clinical Microbiology. 42 (8): 3711–30. doi:10.1128/JCM.42.8.3711-3730.2004. PMC 497648
 . PMID 15297521.
. PMID 15297521. - ↑ Achtman, M.; Zurth, K.; Morelli, G.; Torrea, G.; Guiyoule, A.; Carniel, E. (1999). "Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis". Proceedings of the National Academy of Sciences of the United States of America. 96 (24): 14043–14048. Bibcode:1999PNAS...9614043A. doi:10.1073/pnas.96.24.14043. PMC 24187
 . PMID 10570195.
. PMID 10570195. - ↑ Young, J. M.; Park, D. -C. (2007). "Probable synonymy of the nitrogen-fixing genus Azotobacter and the genus Pseudomonas". International Journal of Systematic and Evolutionary Microbiology. 57 (12): 2894–2901. doi:10.1099/ijs.0.64969-0. PMID 18048745.
- ↑ Rediers, H; Vanderleyden, J; De Mot, R (2004). "Azotobacter vinelandii: a Pseudomonas in disguise?". Microbiology (Reading, England). 150 (Pt 5): 1117–9. doi:10.1099/mic.0.27096-0. PMID 15133068.
- ↑ Xu, D; Côté, JC (2003). "Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 3' end 16S rDNA and 5' end 16S-23S ITS nucleotide sequences". International Journal of Systematic and Evolutionary Microbiology. 53 (Pt 3): 695–704. doi:10.1099/ijs.0.02346-0. PMID 12807189.
- ↑ Young, J.; Kuykendall, L.; Martínez-Romero, E.; Kerr, A.; Sawada, H. (2001). "A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. Rhizogenes, R. Rubi, R. Undicola and R. Vitis". International Journal of Systematic and Evolutionary Microbiology. 51 (Pt 1): 89–103. doi:10.1099/00207713-51-1-89. PMID 11211278.
- ↑ Farrand, S.; Van Berkum, P.; Oger, P. (2003). "Agrobacterium is a definable genus of the family Rhizobiaceae". International Journal of Systematic and Evolutionary Microbiology. 53 (Pt 5): 1681–1687. doi:10.1099/ijs.0.02445-0. PMID 13130068.
- ↑ Young, J.; Kuykendall, L.; Martínez-Romero, E.; Kerr, A.; Sawada, H. (2003). "Classification and nomenclature of Agrobacterium and Rhizobium". International Journal of Systematic and Evolutionary Microbiology. 53 (Pt 5): 1689–1695. doi:10.1099/ijs.0.02762-0. PMID 13130069.
- ↑ C. Jeffrey. 1989. Biological Nomenclature, 3rd ed. Edward Arnold, London, 86 pp.
- ↑ app. 9 of BC http://www.ncbi.nlm.nih.gov/books/NBK8808/
- 1 2 3 4 Help! Latin! How to avoid the most common mistakes while giving Latin names to newly discovered prokaryotes. Microbiología (Sociedad Española de Microbiología), 1996, 12, 473-475. http://www.bacterio.cict.fr/trueper.html
- ↑ Classification of Phyla entry in LPSN [Euzéby, J.P. (1997). "List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet". Int J Syst Bacteriol. 47 (2): 590–2. doi:10.1099/00207713-47-2-590. ISSN 0020-7713. PMID 9103655.]
- ↑ STACKEBRANDT, E.; RAINEY; WARD-RAINEY, N.L. (1997). "Proposal for a new hierarchic classification system, Actinobacteria classis nov". Int. J. Syst. Bacteriol. 47: 479–491. doi:10.1099/00207713-47-2-479.
- 1 2 Classification entry in LPSN [Euzéby, J.P. (1997). "List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet". Int J Syst Bacteriol. 47 (2): 590–2. doi:10.1099/00207713-47-2-590. ISSN 0020-7713. PMID 9103655.]
- 1 2 Hierarchical classification of Prokaryotes entry in LPSN [Euzéby, J.P. (1997). "List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet". Int J Syst Bacteriol. 47 (2): 590–2. doi:10.1099/00207713-47-2-590. ISSN 0020-7713. PMID 9103655.]
- ↑ Murray, R. G. E. 1984. The higher taxa, or, a place for everything . . .?, p. 33. In N. R. Krieg and J. G. Holt (ed.), Bergey’s manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore.
- ↑ SCHROETER (J.). In: F. COHN (ed.), Kryptogamenflora von Schlesien. Band 3, Heft 3, Pilze. J.U. Kern's Verlag, Breslau, 1885-1889, pp. 1-814.
- ↑ Salmonella entry in LPSN [Euzéby, J.P. (1997). "List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet". Int J Syst Bacteriol. 47 (2): 590–2. doi:10.1099/00207713-47-2-590. ISSN 0020-7713. PMID 9103655.]
- ↑ R. E. BUCHANAN, Taxonomy, Annu. Rev. Microbiol. 1955.9:1-20. http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.09.100155.000245
- ↑ ftp://ftp.ncbi.nih.gov/pub/taxonomy/