Actinobacteria
| Actinobacteria | |
|---|---|
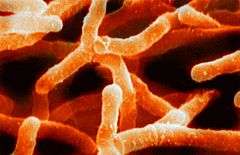 | |
| Scanning electron micrograph of Actinomyces israelii. | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Actinobacteria Goodfellow, 2012 |
| Classes | |
| Synonyms | |
| |
The Actinobacteria are a phylum of Gram-positive bacteria. They can be terrestrial or aquatic.[1] They are of great economic importance to humans because agriculture and forests depend on their contributions to soil systems. In soil, they behave much like fungi, helping to decompose the organic matter of dead organisms so the molecules can be taken up anew by plants. In this role the colonies often grow extensive mycelia, like a fungus would, and the name of an important order of the phylum, Actinomycetales (the actinomycetes), reflects that they were long believed to be fungi. Some soil actinobacteria (such as Frankia) live symbiotically with the plants whose roots pervade the soil, fixing nitrogen for the plants in exchange for access to some of the plant's saccharides.
Beyond the great interest in Actinobacteria for their soil role, much is yet to be learned about them. Although currently understood primarily as soil bacteria, they might be more abundant in fresh waters.[2] Actinobacteria is one of the dominant bacterial phyla and contains one of the largest of bacterial genera, Streptomyces.[3] Streptomyces and other actinobacteria are major contributors to biological buffering of soils.[4] They are also the source of many antibiotics.
Although some of the largest and most complex bacterial cells belong to the Actinobacteria, the group of marine Actinomarinales has been described as possessing the smallest free-living prokaryotic cells.[5]
General
Most Actinobacteria of medical or economic significance are in subclass Actinobacteridae, and belong to the order Actinomycetales. While many of these cause disease in humans, Streptomyces is notable as a source of antibiotics.
Of those Actinobacteria not in the Actinomycetales, Gardnerella is one of the most researched. Classification of Gardnerella is controversial, and MeSH catalogues it as both a Gram-positive and Gram-negative organism.[6]
Actinobacteria, especially Streptomyces spp., are recognized as the producers of many bioactive metabolites that are useful to humans in medicine, such as antibacterials,[7] antifungals,[8] antivirals, antithrombotics, immunomodifiers, antitumor drugs, and enzyme inhibitors; and in agriculture, including insecticides, herbicides, fungicides, and growth-promoting substances for plants and animals.[9][10] Actinobacteria-derived antibiotics that are important in medicine include aminoglycosides, anthracyclines, chloramphenicol, macrolide, tetracyclines etc.
Actinobacteria have high guanine and cytosine content in their DNA.[11] The G+C content of Actinobacteria can be as high as 70%, though some may have a low G+C content.[12]
Analysis of glutamine synthetase sequence has been suggested for phylogenetic analysis of the Actinobacteria.[13]
Phylogeny
The phylogeny is based on 16S rRNA-based LTP release 123 by 'The All-Species Living Tree' Project.[14]
| |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |
Taxonomy
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN)[15] and National Center for Biotechnology Information (NCBI).[16]
- Species "Cathayosporangium alboflavum" ♠ Runmao et al. 1995
- Species "Candidatus Planktophila limnetica" Jezbera et al. 2009
- Species "Tonsillophilus suis" ♠ Azuma and Bak 1980
- Class Rubrobacteria Suzuki 2013 (Rubrobacteridae Rainey et al. 1997 emend. Zhi et al. 2009)
- Order Rubrobacterales Rainey et al. 1997 emend. Zhi et al. 2009
- Family Rubrobacteraceae Rainey et al. 1997
- Order Rubrobacterales Rainey et al. 1997 emend. Zhi et al. 2009
- Class Thermoleophilia Suzuki & Whitman 2013 (Thermoleophilidae)
- Order Gaiellales Albuquerque et al. 2012
- Family Gaiellaceae Albuquerque et al. 2012
- Order Thermoleophilales Reddy & Garcia-Pichel 2009
- Family Thermoleophilaceae Stackebrandt 2005 emend. Zhi, Li & Stackebrandt 2009
- Order Solirubrobacterales Reddy & Garcia-Pichel 2009
- Family Conexibacteraceae Stackebrandt 2005 emend. Zhi, Li & Stackebrandt 2009
- Family Patulibacteraceae Takahashi et al. 2006 emend. Zhi, Li & Stackebrandt 2009
- Family Solirubrobacteraceae Stackebrandt 2005 emend. Zhi et al. 2009
- Order Gaiellales Albuquerque et al. 2012
- Class Coriobacteriia Konig 2013 emend. Gupta et al. 2013 (Coriobacteridae Stackebrandt et al. 1997 emend. Zhi et al. 2009)
- Order Coriobacteriales Stackebrandt et al. 1997 emend. Gupta et al. 2013
- Family Atopobiaceae Gupta et al. 2013
- Family Coriobacteriaceae Stackebrandt, Rainey & Ward-Rainey 1997 emend. Zhi, Li & Stackebrandt 2009
- Order Eggerthellales Gupta et al. 2013
- Family Eggerthellaceae Gupta et al. 2013
- Order Coriobacteriales Stackebrandt et al. 1997 emend. Gupta et al. 2013
- Class Acidimicrobiia Norris 2013 (Acidimicrobidae Stackebrandt et al. 1997 emend. Zhi et al. 2009)
- Order Acidimicrobiales Stackebrandt et al. 1997 emend. Zhi et al. 2009 [Acidimicrobineae Garrity, Bell & Lilburn 2003]
- Family Acidimicrobiaceae Stackebrandt et al. 1997 emend. Zhi et al. 2009
- Family Iamiaceae Kurahashi et al. 2009
- Family "Microthrixaceae" Joseph et al. 2003
- Order Acidimicrobiales Stackebrandt et al. 1997 emend. Zhi et al. 2009 [Acidimicrobineae Garrity, Bell & Lilburn 2003]
- Class Nitriliruptoria Ludwig et al. 2013 (Nitriliruptoridae Kurahashi et al. 2010)
- Order Egibacterales Zhang et al. 2016
- Family Egibacteraceae Zhang et al. 2016
- Order Egicoccales Zhang et al. 2015
- Family Egicoccaceae Zhang et al. 2015
- Order Euzebyales Kurahashi et al. 2010
- Family Euzebyaceae Kurahashi et al. 2010
- Order Nitriliruptorales Sorokin et al. 2009
- Family Nitriliruptoraceae Sorokin et al. 2009
- Order Egibacterales Zhang et al. 2016
- Class Actinobacteria Stackebrandt et al. 1997 (Actinobacteridae Stackebrandt et al. 1997 emend. Zhi et al. 2009)
- Species ?"Boyliae praeputiale" ♠ Yates et al. 2002
- Order ?"Actinomarinales" Ghai et al. 2013 ["Actinomarinidae" Ghai et al. 2013]
- Family "Actinomarinaceae" Ghai et al. 2013
- Order Acidothermales Sen et al. 2014
- Family Acidothermaceae Rainey, Ward-Rainey & Stackebrandt 1997 emend. Zhi et al. 2009
- Order "Catenulisporales" Donadio et al 2012 (Catenulisporineae Cavaletti et al. 2006 emend. Zhi et al. 2009)
- Family Actinospicaceae Cavaletti et al. 2006 emend. Zhi et al. 2009
- Family Catenulisporaceae Busti et al. 2006 emend. Zhi et al. 2009
- Order "Streptomycetales" Kampfer 2012 (Streptomycetineae Rainey et al. 1997 emend. Zhi et al. 2009)
- Family Streptomycetaceae Waksman & Henrici 1943 emend. Zhi et al. 2009
- Order Geodermatophilales Sen et al. 2014
- Family Geodermatophilaceae Normand 2006 emend. Zhi, Li & Stackebrandt 2009
- Order Frankiales Sen et al. 2014 (Frankineae Stackebrandt, Rainey & Ward-Rainey 1997 emend. Zhi, Li & Stackebrandt 2009)
- Family Motilibacteraceae Lee 2013
- Family Cryptosporangiaceae Zhi, Li & Stackebrandt 2009
- Family Frankiaceae Becking 1970 emend. Zhi et al. 2009
- Family Sporichthyaceae Rainey et al. 1997 emend. Zhi, Li & Stackebrandt 2009
- Order Nakamurellales Sen et al. 2014
- Family Nakamurellaceae Tao et al. 2004 emend. Zhi, Li & Stackebrandt 2009 [Microsphaeraceae Rainey et al. 1997]
- Order "Kineosporiales" Kampfer 2012 (Kineosporineae Zhi, Li & Stackebrandt 2009)
- Family Kineosporiaceae Zhi, Li & Stackebrandt 2009
- Order Micrococcales Prevot 1940 [Arthrobacteria; Micrococcineae Stackebrandt et al. 1997 emend. Yassin et al. 2011; incl. Flavobacterium oceanosedimentum; Bifidobacteriales Stackebrandt et al. 1997 emend. Zhi et al. 2009; Actinomycetales Buchanan 1917 emend. Zhi et al. 2009]
- Family Intrasporangiaceae
- Family Microbacteriaceae
- Family Cellulomonadceae
- Family Promicromonosporaceae
- Family Sanguibacteraceae
- Family Rarobacteraceae
- Family Jonesiaceae
- Family Bogoriellaceae
- Family Beutenbergiaceae
- Family Ruaniaceae
- Family Actinomycetaceae
- Family Demequinaceae
- Family Dermabacteraceae
- Family Brevibacteriaceae
- Family Micrococcaceae {incl. Bifidobacteriaceae]])
- Order "Propionibacteriales" Patrick & McDowell 2012 (Propionibacterineae Rainey et al. 1997 emend. Zhi et al. 2009)
- Family Nocardioidaceae
- Family Propionibacteriaceae
- Order "Streptosporangiales" Goodfellow 2012 (Streptosporangineae Ward-Rainey et al. 1997 emend. Zhi, Li & Stackebrandt 2009)
- Family Streptosporangiaceae
- Family Nocardiopsaceae
- Family Thermomonosporaceae
- Order "Jiangellales" Tang et al. 2012 (Jiangellineae Tang et al. 2011)
- Family Jiangellaceae Tang et al. 2011
- Order "Glycomycetales" Labeda 2012 (Glycomycetineae Rainey, Ward-Rainey & Stackebrandt 1997 emend. Zhi, Li & Stackebrandt 2009)
- Family Glycomycetaceae Rainey, Ward-Rainey & Stackebrandt 1997 emend. Zhi, Li & Stackebrandt 2009
- Order "Micromonosporales" Genilloud 2012 (Micromonosporineae Stackebrandt et al. 1997 emend. Zhi et al. 2009 ; Actinoplanales]
- Family Micromonosporaceae
- Order "Pseudonocardiales" Labeda & Goodfellow 2012 (Pseudonocardineae Stackebrandt et al. 1997 emend. Labeda et al. 2011)
- Family Pseudonocardiaceae
- Order "Corynebacteriales" Goodfellow & Jones 2012 (Corynebacterineae Stackebrandt et al. 1997 emend. Zhi et al. 2009; Mycobacteria)
- Family Segniliparaceae
- Family Nocardiaceae [Gordoniaceae; Williamsiaceae]
- Family Dietziaceae
- Family Corynebacteriaceae
- Family Tsukamurellaceae
- Family Mycobacteriaceae
Notes:
♠ Strains found at the National Center for Biotechnology Information (NCBI) but not listed in the List of Prokaryotic names with Standing in Nomenclature (LSPN)
See also
References
- ↑ Servin JA, Herbold CW, Skophammer RG, Lake JA (January 2008). "Evidence excluding the root of the tree of life from the actinobacteria". Mol. Biol. Evol. 25 (1): 1–4. doi:10.1093/molbev/msm249. PMID 18003601.
- ↑ Ghai R, Rodriguez-Valera F, McMahon KD, et al. (2011). Lopez-Garcia P, ed. "Metagenomics of the water column in the pristine upper course of the Amazon river". PLOS ONE. 6 (8): e23785. doi:10.1371/journal.pone.0023785. PMC 3158796
 . PMID 21915244.
. PMID 21915244. - ↑ C.Michael Hogan. 2010. Bacteria. Encyclopedia of Earth. eds. Sidney Draggan and C.J.Cleveland, National Council for Science and the Environment, Washington DC
- ↑ Ningthoujam, Debananda S.; Sanasam, Suchitra; Tamreihao, K; Nimaichand, Salam (November 2009). "Antagonistic activities of local actinomycete isolates against rice fungal pathogens". African Journal of Microbiology Research. 3 (11): 737–742.
- ↑ Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F (2013). "Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria". Scientific Reports. 3: 2471. doi:10.1038/srep02471. PMC 3747508
 . PMID 23959135.
. PMID 23959135.
- ↑ Gardnerella at the US National Library of Medicine Medical Subject Headings (MeSH)
- ↑ Mahajan, GB (2012). "Antibacterial agents from actinomycetes - a review". Frontiers in Bioscience. 4: 240–53. doi:10.2741/e373.
- ↑ Gupte, M.; Kulkarni, P.; Ganguli, B.N. (2002). "Antifungal Antibiotics". Appl. Microbiol. Biotechnol. 58: 46–57.
- ↑ Bressan, W (2003). "Biological control of maize seed pathogenic fungi by use of actinomycetes". Biocontrol. 48 (2): 233–240. doi:10.1023/a:1022673226324.
- ↑ Atta, M.A (2009). "Antimycin-A Antibiotic Biosynthesis Produced by Streptomyces Sp. AZ-AR-262: Taxonomy, Fermentation, Purification and Biological Activities". Austral. J. Basic and Appl. Sci. 3: 126–135.
- ↑ Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G. F.; Chater, K. F.; van Sinderen, D. (5 September 2007). "Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum". Microbiology and Molecular Biology Reviews. 71 (3): 495–548. doi:10.1128/MMBR.00005-07. PMC 2168647
 . PMID 17804669.
. PMID 17804669. - ↑ Ghai R, McMahon KD, Rodriguez-Valera F (2012). "Breaking a paradigm:cosmopolitan and abundant freshwater actinobacteria are low GC". Environmental Microbiology Reports. 4 (1): 29–35. doi:10.1111/j.1758-2229.2011.00274.x. PMID 23757226.
- ↑ Hayward D, van Helden PD, Wiid IJ (2009). "Glutamine synthetase sequence evolution in the mycobacteria and their use as molecular markers for Actinobacteria speciation". BMC Evol. Biol. 9: 48. doi:10.1186/1471-2148-9-48. PMC 2667176
 . PMID 19245690.
. PMID 19245690. - ↑ 'The All-Species Living Tree' Project."16S rRNA-based LTP release 123 (full tree)" (PDF). Silva Comprehensive Ribosomal RNA Database. Retrieved 2016-03-20.
- ↑ J.P. Euzéby. "Actinobacteria". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 2016-03-20.
- ↑ Sayers; et al. "Actinobacteria". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2016-03-20.
Further reading
- Pandey, B.; Ghimire, P.; Agrawal, V.P. (January 12–15, 2004). Studies on the antibacterial activity of the Actinomycetes isolated from the Khumbu Region of Nepal (PDF). International Conference on the Great Himalayas: Climate, Health, Ecology, Management and Conservation. Kathmandu.
- Baltz, R.H. (2005). "Antibiotic discovery from actinomycetes: Will a renaissance follow the decline and fall?". SIM News. 55: 186–196.
- Baltz, R.H. (2007). "Antimicrobials from Actinomycetes: Back to the Future". Microbe. 2 (3): 125–131.
External links
| Wikispecies has information related to: Actinobacteria |